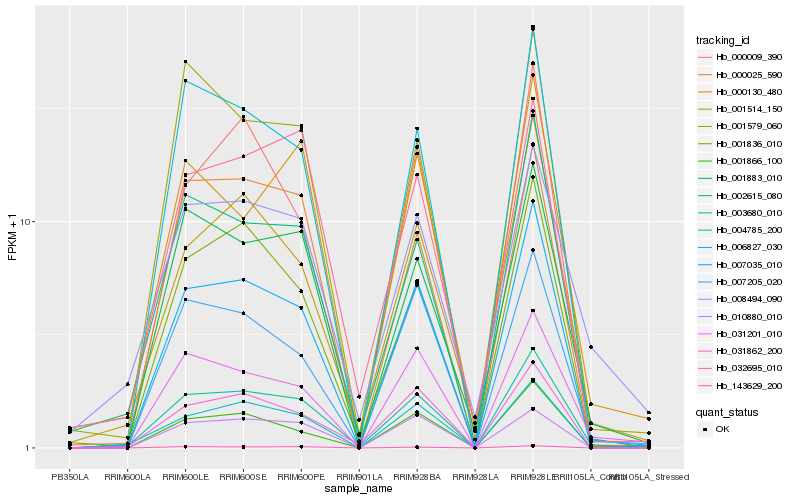
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003680_010 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_007035_010 |
0.0867141058 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002615_080 |
0.1017078122 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 4 |
Hb_004785_200 |
0.1019293596 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 5 |
Hb_031862_200 |
0.1028597174 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 6 |
Hb_006827_030 |
0.1051513078 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 7 |
Hb_001883_010 |
0.1063513025 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 8 |
Hb_001836_010 |
0.1115811629 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 9 |
Hb_032695_010 |
0.1207555627 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 10 |
Hb_001514_150 |
0.1208599772 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 11 |
Hb_031201_010 |
0.1210275182 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001866_100 |
0.1217556533 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 13 |
Hb_143629_200 |
0.1235867171 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 14 |
Hb_007205_020 |
0.1242120146 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 15 |
Hb_000025_590 |
0.1268683331 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_008494_090 |
0.1294055542 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 17 |
Hb_001579_060 |
0.1303764844 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 18 |
Hb_010880_010 |
0.1339091338 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000130_480 |
0.135155877 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 20 |
Hb_000009_390 |
0.1368130419 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |