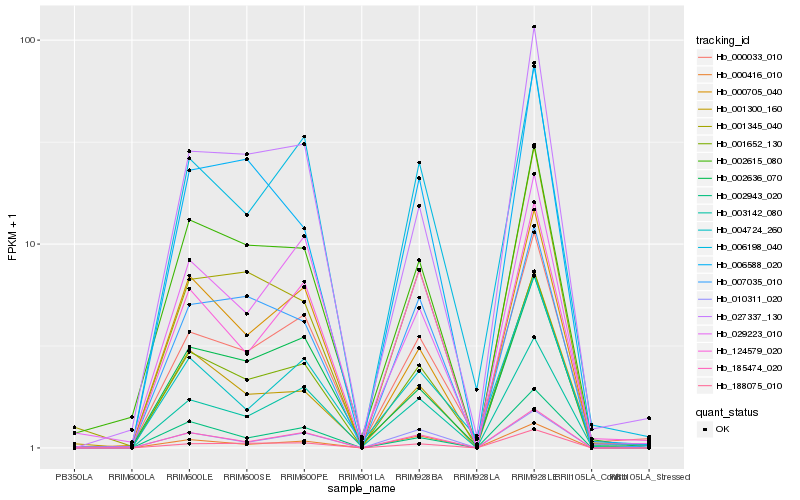
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001652_130 |
0.0 |
- |
- |
hypothetical protein B456_002G193900 [Gossypium raimondii] |
| 2 |
Hb_185474_020 |
0.0832659883 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 3 |
Hb_188075_010 |
0.0852635012 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 4 |
Hb_000033_010 |
0.0895789056 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 5 |
Hb_006198_040 |
0.0972558383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003142_080 |
0.1026894759 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 7 |
Hb_002943_020 |
0.1071688501 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 8 |
Hb_002615_080 |
0.1089598334 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 9 |
Hb_027337_130 |
0.1096626654 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 10 |
Hb_004724_260 |
0.113063147 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 11 |
Hb_010311_020 |
0.1178698628 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000416_010 |
0.118812884 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 13 |
Hb_001345_040 |
0.1193784646 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 14 |
Hb_000705_040 |
0.1217600649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006588_020 |
0.1252157221 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_007035_010 |
0.1263088361 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002636_070 |
0.1280394188 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 18 |
Hb_124579_020 |
0.1283693009 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 19 |
Hb_001300_160 |
0.1295331551 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 20 |
Hb_029223_010 |
0.1300378436 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |