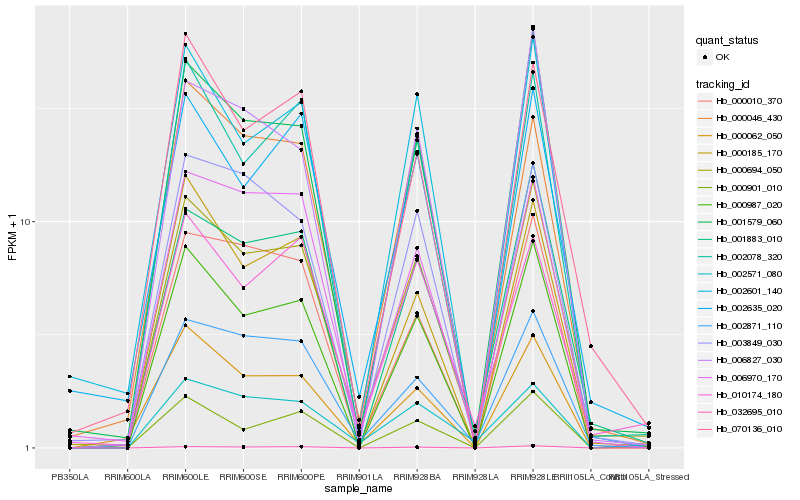
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000694_050 |
0.0 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 2 |
Hb_001579_060 |
0.0702800483 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 3 |
Hb_001883_010 |
0.0830019249 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 4 |
Hb_000987_020 |
0.0849779486 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 5 |
Hb_002871_110 |
0.0990275516 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 6 |
Hb_002601_140 |
0.0996247773 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 7 |
Hb_002078_320 |
0.1040583193 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_002635_020 |
0.1056102525 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 9 |
Hb_032695_010 |
0.1107548585 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 10 |
Hb_000901_010 |
0.1118703853 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 11 |
Hb_003849_030 |
0.1126564233 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 12 |
Hb_070136_010 |
0.1147086962 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 13 |
Hb_006827_030 |
0.1158631499 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 14 |
Hb_000062_050 |
0.1159025398 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_006970_170 |
0.1159476021 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000010_370 |
0.1166946516 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 17 |
Hb_010174_180 |
0.1175260706 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_000046_430 |
0.1184621372 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 19 |
Hb_000185_170 |
0.1186186485 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_002571_080 |
0.1197388167 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |