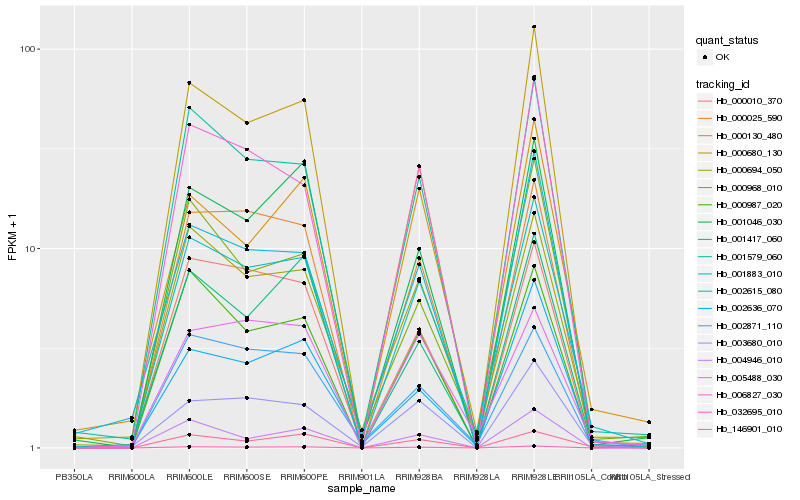
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001883_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 2 |
Hb_001579_060 |
0.0795132167 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 3 |
Hb_032695_010 |
0.0810957592 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 4 |
Hb_000694_050 |
0.0830019249 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 5 |
Hb_000680_130 |
0.0966056532 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 6 |
Hb_000025_590 |
0.0996421909 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_006827_030 |
0.1009473903 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 8 |
Hb_003680_010 |
0.1063513025 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000987_020 |
0.1065448662 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 10 |
Hb_002871_110 |
0.111075701 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 11 |
Hb_000010_370 |
0.1166667418 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 12 |
Hb_146901_010 |
0.1198626064 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 13 |
Hb_002615_080 |
0.1204155576 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 14 |
Hb_001417_060 |
0.1236548836 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 15 |
Hb_001046_030 |
0.124619292 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_004946_010 |
0.1254592592 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 17 |
Hb_002636_070 |
0.1260965966 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 18 |
Hb_005488_030 |
0.1266484975 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 19 |
Hb_000130_480 |
0.1268109826 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 20 |
Hb_000968_010 |
0.1280347156 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |