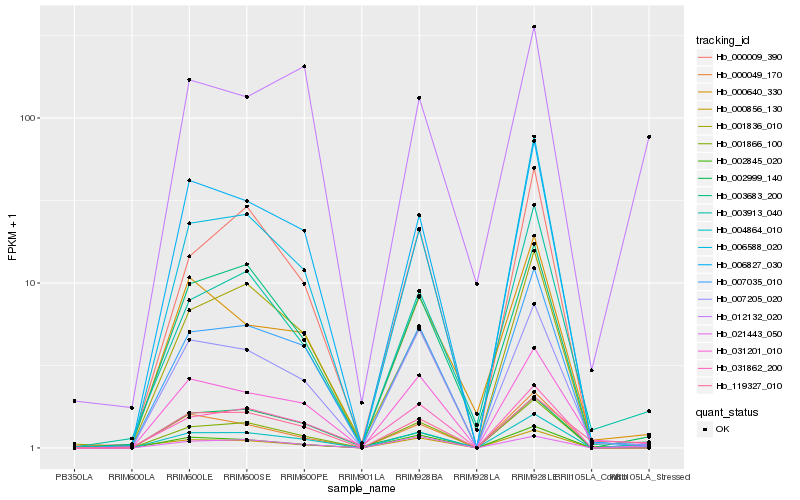
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004864_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10003960mg, partial [Citrus clementina] |
| 2 |
Hb_021443_050 |
0.1349452001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007035_010 |
0.1404947708 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_031201_010 |
0.1440007062 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_001866_100 |
0.1468977624 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 6 |
Hb_000856_130 |
0.147699442 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_007205_020 |
0.1511549783 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 8 |
Hb_003913_040 |
0.1512262994 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 9 |
Hb_119327_010 |
0.153168209 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 10 |
Hb_006588_020 |
0.1586680166 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_006827_030 |
0.160586265 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 12 |
Hb_002845_020 |
0.1640138274 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 13 |
Hb_002999_140 |
0.1643085833 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 14 |
Hb_000049_170 |
0.1663328844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000009_390 |
0.1694035362 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000640_330 |
0.1697818591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001836_010 |
0.1733368478 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 18 |
Hb_012132_020 |
0.1734651908 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 19 |
Hb_003683_200 |
0.1745548875 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 20 |
Hb_031862_200 |
0.177963856 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |