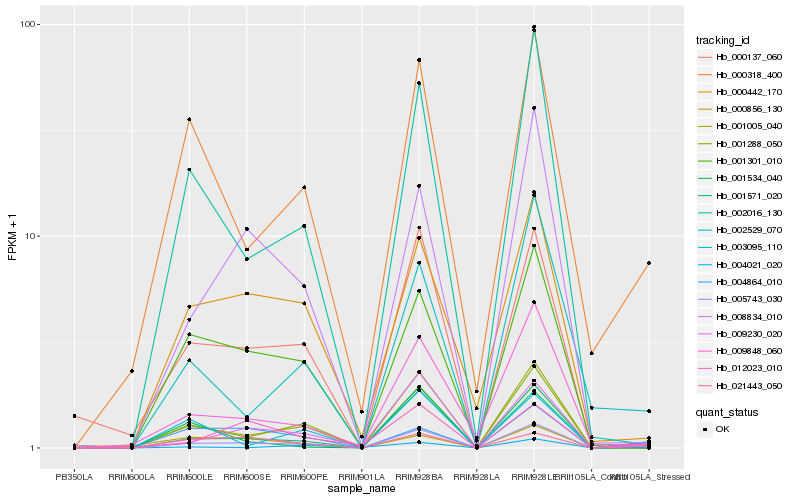
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005743_030 |
0.0 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 2 |
Hb_000318_400 |
0.183004678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001005_040 |
0.2018130076 |
- |
- |
- |
| 4 |
Hb_004864_010 |
0.2061950971 |
- |
- |
hypothetical protein CICLE_v10003960mg, partial [Citrus clementina] |
| 5 |
Hb_012023_010 |
0.2062908785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_021443_050 |
0.206689366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001534_040 |
0.2083919887 |
- |
- |
hypothetical protein JCGZ_08436 [Jatropha curcas] |
| 8 |
Hb_004021_020 |
0.2089370528 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 9 |
Hb_001288_050 |
0.2106792536 |
- |
- |
- |
| 10 |
Hb_001571_020 |
0.2124638254 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850-like [Citrus sinensis] |
| 11 |
Hb_002016_130 |
0.2134820668 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 12 |
Hb_000856_130 |
0.2142882566 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_009848_060 |
0.2144422383 |
- |
- |
unknown [Glycine max] |
| 14 |
Hb_002529_070 |
0.217501043 |
- |
- |
PREDICTED: putative late blight resistance protein homolog R1B-19 [Pyrus x bretschneideri] |
| 15 |
Hb_001301_010 |
0.2178973041 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_009230_020 |
0.2212751685 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000137_060 |
0.2217584753 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 18 |
Hb_000442_170 |
0.2230547354 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_008834_010 |
0.2235368248 |
- |
- |
hypothetical protein POPTR_0016s04660g [Populus trichocarpa] |
| 20 |
Hb_003095_110 |
0.2251676006 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |