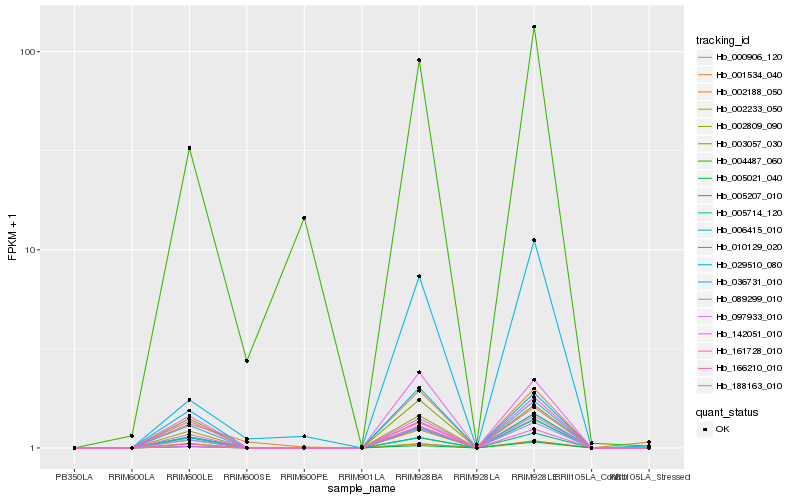
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089299_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000906_120 |
0.0643518041 |
- |
- |
hypothetical protein VITISV_006406 [Vitis vinifera] |
| 3 |
Hb_006415_010 |
0.0691780386 |
- |
- |
- |
| 4 |
Hb_142051_010 |
0.0743467545 |
- |
- |
- |
| 5 |
Hb_002188_050 |
0.0991247053 |
- |
- |
PREDICTED: serine carboxypeptidase-like 2 [Jatropha curcas] |
| 6 |
Hb_002809_090 |
0.1035415136 |
- |
- |
- |
| 7 |
Hb_029510_080 |
0.1186927848 |
- |
- |
- |
| 8 |
Hb_003057_030 |
0.1236333346 |
- |
- |
hypothetical protein JCGZ_06668 [Jatropha curcas] |
| 9 |
Hb_005021_040 |
0.1356226453 |
- |
- |
PREDICTED: uncharacterized protein LOC103848240 [Brassica rapa] |
| 10 |
Hb_161728_010 |
0.1477883952 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 11 |
Hb_188163_010 |
0.1480198637 |
- |
- |
hypothetical protein CISIN_1g048732mg [Citrus sinensis] |
| 12 |
Hb_002233_050 |
0.1714042082 |
- |
- |
PREDICTED: uncharacterized protein LOC104879391 [Vitis vinifera] |
| 13 |
Hb_010129_020 |
0.1720741519 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_004487_060 |
0.1752034662 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_005207_010 |
0.1758721415 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Jatropha curcas] |
| 16 |
Hb_005714_120 |
0.1791750505 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_036731_010 |
0.183340759 |
- |
- |
- |
| 18 |
Hb_001534_040 |
0.1835229569 |
- |
- |
hypothetical protein JCGZ_08436 [Jatropha curcas] |
| 19 |
Hb_097933_010 |
0.1881273111 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_166210_010 |
0.1893564509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |