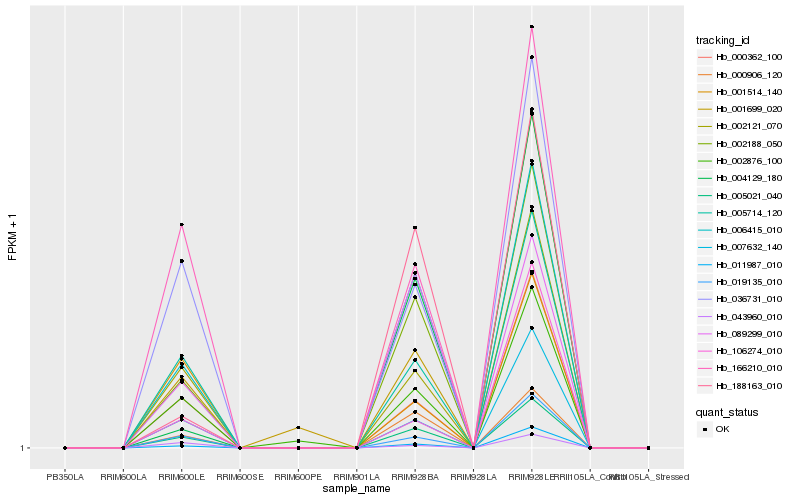
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005021_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103848240 [Brassica rapa] |
| 2 |
Hb_002188_050 |
0.0415203484 |
- |
- |
PREDICTED: serine carboxypeptidase-like 2 [Jatropha curcas] |
| 3 |
Hb_006415_010 |
0.0671946305 |
- |
- |
- |
| 4 |
Hb_000906_120 |
0.0820710582 |
- |
- |
hypothetical protein VITISV_006406 [Vitis vinifera] |
| 5 |
Hb_002121_070 |
0.0824750719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005714_120 |
0.0892964334 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000362_100 |
0.0897589538 |
- |
- |
- |
| 8 |
Hb_007632_140 |
0.0975901758 |
- |
- |
- |
| 9 |
Hb_036731_010 |
0.1116468713 |
- |
- |
- |
| 10 |
Hb_019135_010 |
0.1128896151 |
- |
- |
- |
| 11 |
Hb_002876_100 |
0.1220328788 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |
| 12 |
Hb_166210_010 |
0.1283918243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_089299_010 |
0.1356226453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011987_010 |
0.1463055138 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 15 |
Hb_001699_020 |
0.150987185 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 16 |
Hb_188163_010 |
0.1565456917 |
- |
- |
hypothetical protein CISIN_1g048732mg [Citrus sinensis] |
| 17 |
Hb_106274_010 |
0.1674265554 |
- |
- |
hypothetical protein VITISV_008825 [Vitis vinifera] |
| 18 |
Hb_004129_180 |
0.1683252296 |
- |
- |
hypothetical protein JCGZ_11090 [Jatropha curcas] |
| 19 |
Hb_043960_010 |
0.1687573936 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_001514_140 |
0.1714277654 |
- |
- |
- |