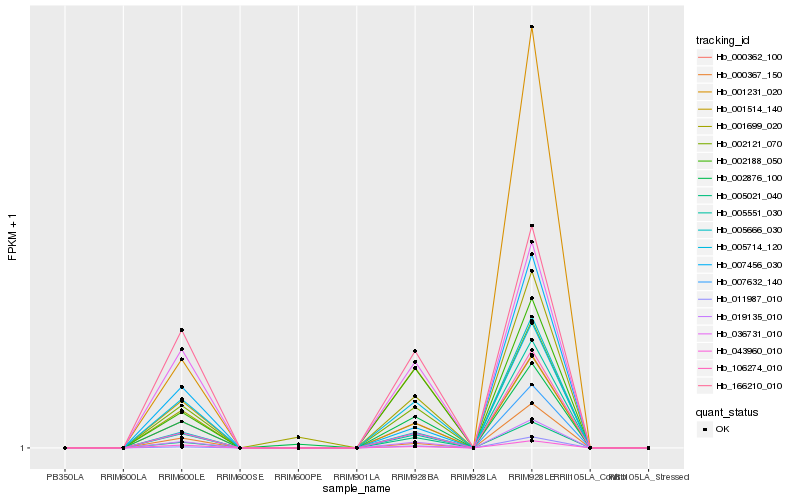
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007632_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_019135_010 |
0.016231346 |
- |
- |
- |
| 3 |
Hb_000362_100 |
0.027966725 |
- |
- |
- |
| 4 |
Hb_002121_070 |
0.0565932046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005714_120 |
0.0896330576 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_106274_010 |
0.0897633515 |
- |
- |
hypothetical protein VITISV_008825 [Vitis vinifera] |
| 7 |
Hb_005021_040 |
0.0975901758 |
- |
- |
PREDICTED: uncharacterized protein LOC103848240 [Brassica rapa] |
| 8 |
Hb_043960_010 |
0.1017577259 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_000367_150 |
0.1061318096 |
- |
- |
- |
| 10 |
Hb_001231_020 |
0.1078026208 |
- |
- |
- |
| 11 |
Hb_011987_010 |
0.1082549441 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 12 |
Hb_036731_010 |
0.1140072033 |
- |
- |
- |
| 13 |
Hb_005551_030 |
0.122210096 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 14 |
Hb_007456_030 |
0.1287552624 |
- |
- |
- |
| 15 |
Hb_166210_010 |
0.1308554133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001514_140 |
0.1309816223 |
- |
- |
- |
| 17 |
Hb_002876_100 |
0.1325340584 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |
| 18 |
Hb_001699_020 |
0.1355774686 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 19 |
Hb_005666_030 |
0.136489622 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |
| 20 |
Hb_002188_050 |
0.1386446771 |
- |
- |
PREDICTED: serine carboxypeptidase-like 2 [Jatropha curcas] |