| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
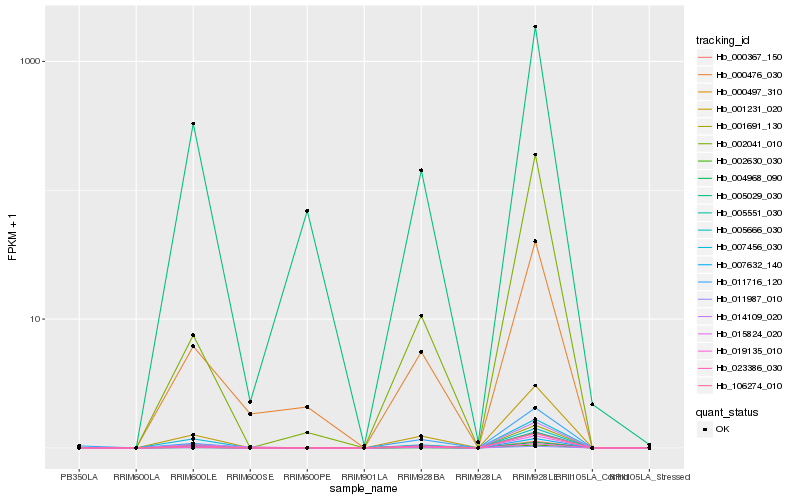
Hb_004968_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104882781 [Vitis vinifera] |
| 2 |
Hb_005666_030 |
0.0062666607 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |
| 3 |
Hb_005551_030 |
0.0175028454 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 4 |
Hb_001231_020 |
0.0323575294 |
- |
- |
- |
| 5 |
Hb_001691_130 |
0.0335315316 |
- |
- |
PREDICTED: uncharacterized protein LOC104587431 [Nelumbo nucifera] |
| 6 |
Hb_106274_010 |
0.0502649452 |
- |
- |
hypothetical protein VITISV_008825 [Vitis vinifera] |
| 7 |
Hb_002630_030 |
0.054527386 |
- |
- |
OSIGBa0132I10.1 [Oryza sativa Indica Group] |
| 8 |
Hb_015824_020 |
0.0710859227 |
- |
- |
- |
| 9 |
Hb_023386_030 |
0.0749326712 |
- |
- |
- |
| 10 |
Hb_011987_010 |
0.0876790772 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 11 |
Hb_000367_150 |
0.0962407282 |
- |
- |
- |
| 12 |
Hb_000497_310 |
0.1042728902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002041_010 |
0.1229768969 |
- |
- |
- |
| 14 |
Hb_019135_010 |
0.1260286314 |
- |
- |
- |
| 15 |
Hb_007456_030 |
0.1336331464 |
- |
- |
- |
| 16 |
Hb_014109_020 |
0.1370268824 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 17 |
Hb_011716_120 |
0.1373864534 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 18 |
Hb_007632_140 |
0.1395060911 |
- |
- |
- |
| 19 |
Hb_000476_030 |
0.1423726547 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 20 |
Hb_005029_030 |
0.1434753346 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 36.4 kDa proline-rich protein-like [Sesamum indicum] |