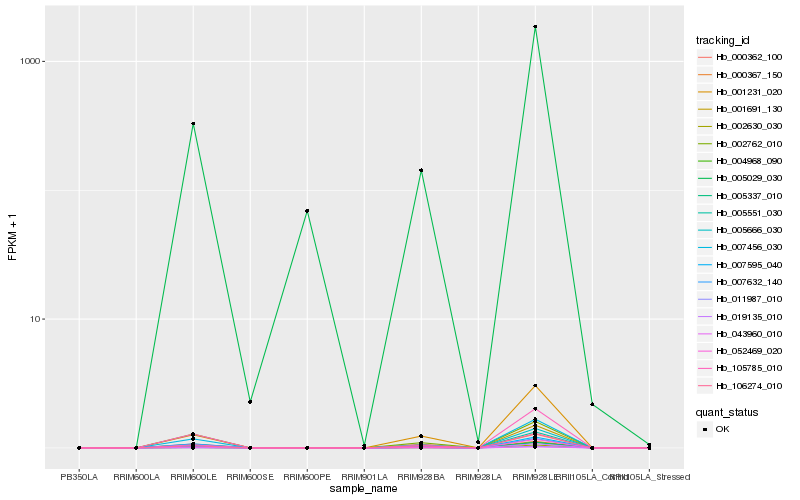
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_007456_030 |
0.0402938323 |
- |
- |
- |
| 3 |
Hb_001691_130 |
0.0719349476 |
- |
- |
PREDICTED: uncharacterized protein LOC104587431 [Nelumbo nucifera] |
| 4 |
Hb_001231_020 |
0.0757509487 |
- |
- |
- |
| 5 |
Hb_106274_010 |
0.0771412096 |
- |
- |
hypothetical protein VITISV_008825 [Vitis vinifera] |
| 6 |
Hb_002630_030 |
0.0806557555 |
- |
- |
OSIGBa0132I10.1 [Oryza sativa Indica Group] |
| 7 |
Hb_005551_030 |
0.0862346994 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 8 |
Hb_105785_010 |
0.0875103473 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 9 |
Hb_019135_010 |
0.0905511967 |
- |
- |
- |
| 10 |
Hb_052469_020 |
0.0959149312 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Vitis vinifera] |
| 11 |
Hb_004968_090 |
0.0962407282 |
- |
- |
PREDICTED: uncharacterized protein LOC104882781 [Vitis vinifera] |
| 12 |
Hb_007595_040 |
0.0973784427 |
- |
- |
ribosomal protein L2 (chloroplast) [Berberis bealei] |
| 13 |
Hb_005666_030 |
0.0984562842 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |
| 14 |
Hb_007632_140 |
0.1061318096 |
- |
- |
- |
| 15 |
Hb_005337_010 |
0.106490399 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 16 |
Hb_043960_010 |
0.1196569987 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_000362_100 |
0.1227539054 |
- |
- |
- |
| 18 |
Hb_005029_030 |
0.1270117742 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 36.4 kDa proline-rich protein-like [Sesamum indicum] |
| 19 |
Hb_011987_010 |
0.1412606041 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 20 |
Hb_002762_010 |
0.1418798899 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |