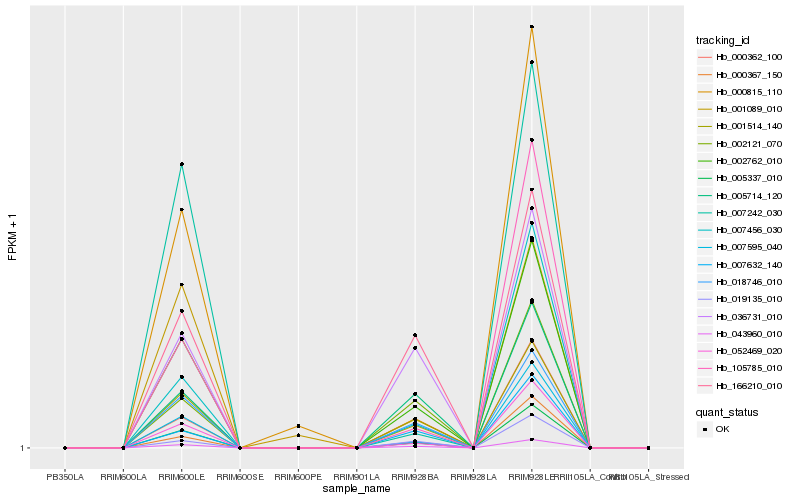
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005337_010 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 2 |
Hb_002762_010 |
0.0505093068 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 3 |
Hb_007595_040 |
0.0554086403 |
- |
- |
ribosomal protein L2 (chloroplast) [Berberis bealei] |
| 4 |
Hb_052469_020 |
0.0574691974 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Vitis vinifera] |
| 5 |
Hb_043960_010 |
0.0637761768 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_007456_030 |
0.0751049701 |
- |
- |
- |
| 7 |
Hb_018746_010 |
0.0974499778 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 8 |
Hb_105785_010 |
0.099334625 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 9 |
Hb_001514_140 |
0.1001788544 |
- |
- |
- |
| 10 |
Hb_000367_150 |
0.106490399 |
- |
- |
- |
| 11 |
Hb_019135_010 |
0.1366030929 |
- |
- |
- |
| 12 |
Hb_000815_110 |
0.1371468446 |
- |
- |
RecName: Full=Casparian strip membrane protein 4; Short=RcCASP4 [Ricinus communis] |
| 13 |
Hb_000362_100 |
0.1384834834 |
- |
- |
- |
| 14 |
Hb_007632_140 |
0.1438318353 |
- |
- |
- |
| 15 |
Hb_002121_070 |
0.1461978126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005714_120 |
0.157858567 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_036731_010 |
0.1588901731 |
- |
- |
- |
| 18 |
Hb_007242_030 |
0.160546965 |
- |
- |
PREDICTED: methylesterase 3-like isoform X2 [Tarenaya hassleriana] |
| 19 |
Hb_166210_010 |
0.1610703844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001089_010 |
0.1717696988 |
- |
- |
PREDICTED: GDSL esterase/lipase 5-like [Jatropha curcas] |