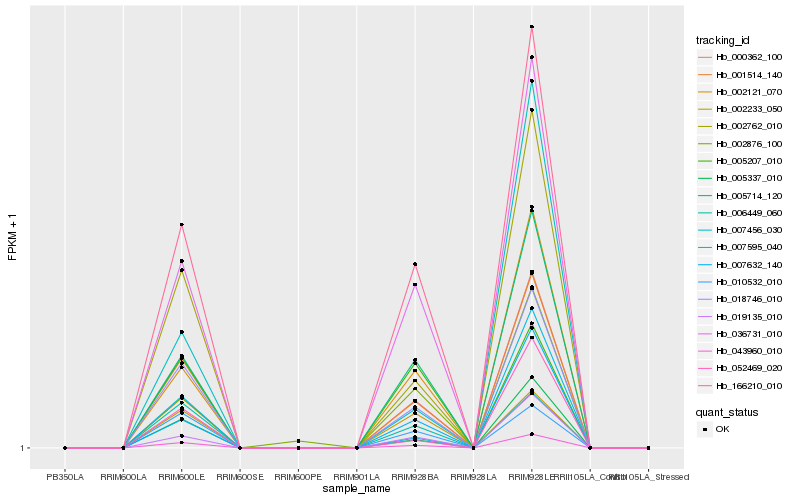
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_018746_010 |
0.0110589284 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 3 |
Hb_043960_010 |
0.0495302886 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_002762_010 |
0.0539302581 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 5 |
Hb_166210_010 |
0.0671826667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_036731_010 |
0.0719880451 |
- |
- |
- |
| 7 |
Hb_005714_120 |
0.0845725089 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_002121_070 |
0.0943254053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005337_010 |
0.1001788544 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 10 |
Hb_010532_010 |
0.1063860738 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 11 |
Hb_006449_060 |
0.1075953572 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 12 |
Hb_000362_100 |
0.1082170731 |
- |
- |
- |
| 13 |
Hb_007632_140 |
0.1309816223 |
- |
- |
- |
| 14 |
Hb_019135_010 |
0.1354311586 |
- |
- |
- |
| 15 |
Hb_002233_050 |
0.1409656521 |
- |
- |
PREDICTED: uncharacterized protein LOC104879391 [Vitis vinifera] |
| 16 |
Hb_007595_040 |
0.1542180111 |
- |
- |
ribosomal protein L2 (chloroplast) [Berberis bealei] |
| 17 |
Hb_007456_030 |
0.1558732919 |
- |
- |
- |
| 18 |
Hb_052469_020 |
0.1561220185 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Vitis vinifera] |
| 19 |
Hb_002876_100 |
0.1585444843 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |
| 20 |
Hb_005207_010 |
0.1607521709 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Jatropha curcas] |