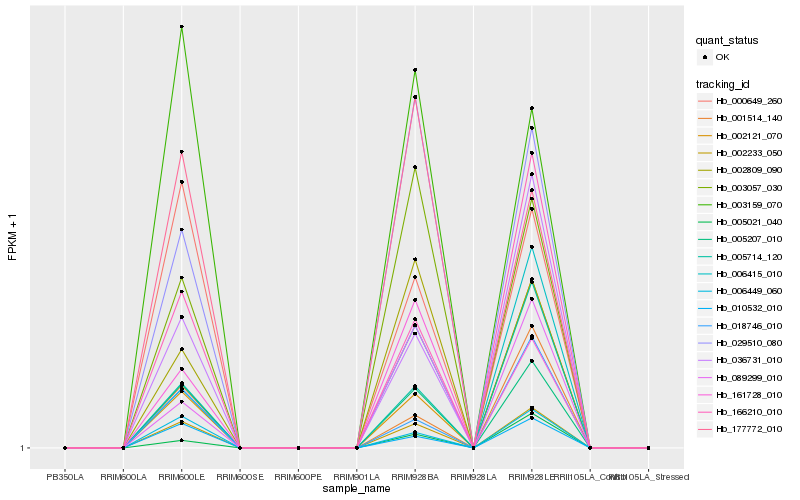
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002233_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104879391 [Vitis vinifera] |
| 2 |
Hb_005207_010 |
0.0222076312 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Jatropha curcas] |
| 3 |
Hb_166210_010 |
0.0888751294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006449_060 |
0.0979250343 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 5 |
Hb_010532_010 |
0.1005203663 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 6 |
Hb_036731_010 |
0.1023731523 |
- |
- |
- |
| 7 |
Hb_000649_260 |
0.1078325845 |
- |
- |
uncharacterized LOC105634108 [Jatropha curcas] |
| 8 |
Hb_005714_120 |
0.1251524435 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_029510_080 |
0.1345125446 |
- |
- |
- |
| 10 |
Hb_003057_030 |
0.1372582466 |
- |
- |
hypothetical protein JCGZ_06668 [Jatropha curcas] |
| 11 |
Hb_001514_140 |
0.1409656521 |
- |
- |
- |
| 12 |
Hb_018746_010 |
0.1446040677 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 13 |
Hb_003159_070 |
0.1446758736 |
- |
- |
Uncharacterized protein TCM_035723 [Theobroma cacao] |
| 14 |
Hb_002809_090 |
0.1468636332 |
- |
- |
- |
| 15 |
Hb_006415_010 |
0.1566076067 |
- |
- |
- |
| 16 |
Hb_002121_070 |
0.1580343455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_177772_010 |
0.1662769032 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X2 [Vitis vinifera] |
| 18 |
Hb_161728_010 |
0.1685635805 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 19 |
Hb_089299_010 |
0.1714042082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005021_040 |
0.1794191248 |
- |
- |
PREDICTED: uncharacterized protein LOC103848240 [Brassica rapa] |