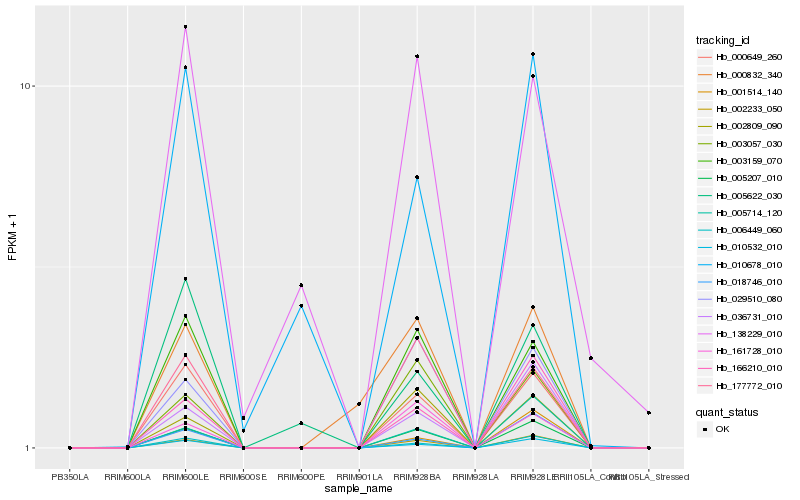
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_260 |
0.0 |
- |
- |
uncharacterized LOC105634108 [Jatropha curcas] |
| 2 |
Hb_006449_060 |
0.0892064226 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 3 |
Hb_010532_010 |
0.0914035841 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 4 |
Hb_003159_070 |
0.0932753702 |
- |
- |
Uncharacterized protein TCM_035723 [Theobroma cacao] |
| 5 |
Hb_005207_010 |
0.0956497671 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Jatropha curcas] |
| 6 |
Hb_002233_050 |
0.1078325845 |
- |
- |
PREDICTED: uncharacterized protein LOC104879391 [Vitis vinifera] |
| 7 |
Hb_177772_010 |
0.1554699828 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X2 [Vitis vinifera] |
| 8 |
Hb_166210_010 |
0.1721016991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005622_030 |
0.177743926 |
- |
- |
transferase, putative [Ricinus communis] |
| 10 |
Hb_010678_010 |
0.1880923057 |
- |
- |
PREDICTED: alpha-glucosidase-like [Jatropha curcas] |
| 11 |
Hb_018746_010 |
0.1896023615 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 12 |
Hb_036731_010 |
0.1898178611 |
- |
- |
- |
| 13 |
Hb_001514_140 |
0.1923236293 |
- |
- |
- |
| 14 |
Hb_003057_030 |
0.1934845187 |
- |
- |
hypothetical protein JCGZ_06668 [Jatropha curcas] |
| 15 |
Hb_029510_080 |
0.193681008 |
- |
- |
- |
| 16 |
Hb_161728_010 |
0.2116513805 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 17 |
Hb_002809_090 |
0.2122534222 |
- |
- |
- |
| 18 |
Hb_005714_120 |
0.2159946621 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000832_340 |
0.2224742757 |
- |
- |
PREDICTED: psbP-like protein 2, chloroplastic isoform X3 [Nelumbo nucifera] |
| 20 |
Hb_138229_010 |
0.2230168598 |
- |
- |
hypothetical protein JCGZ_05736 [Jatropha curcas] |