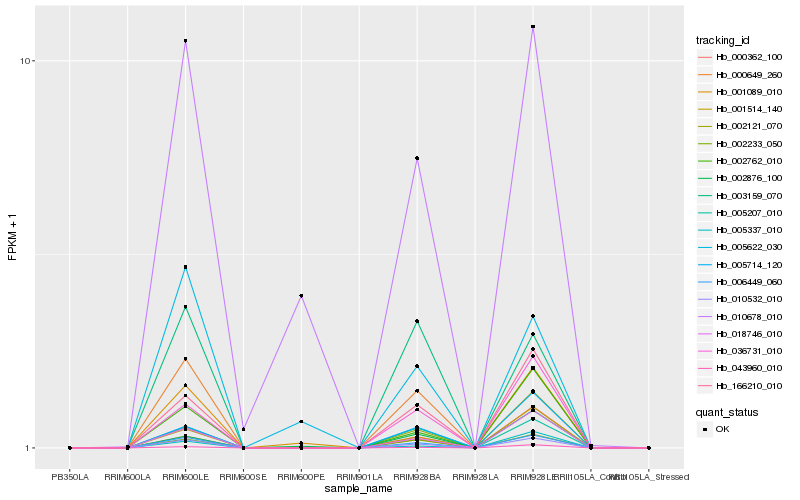
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006449_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 2 |
Hb_010532_010 |
0.0029506 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 3 |
Hb_000649_260 |
0.0892064226 |
- |
- |
uncharacterized LOC105634108 [Jatropha curcas] |
| 4 |
Hb_002233_050 |
0.0979250343 |
- |
- |
PREDICTED: uncharacterized protein LOC104879391 [Vitis vinifera] |
| 5 |
Hb_018746_010 |
0.103195313 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 6 |
Hb_005207_010 |
0.1049919469 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Jatropha curcas] |
| 7 |
Hb_001514_140 |
0.1075953572 |
- |
- |
- |
| 8 |
Hb_166210_010 |
0.1088177186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_036731_010 |
0.1269048054 |
- |
- |
- |
| 10 |
Hb_002762_010 |
0.142559384 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 11 |
Hb_005714_120 |
0.1530336207 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_043960_010 |
0.1560487535 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_010678_010 |
0.1736280049 |
- |
- |
PREDICTED: alpha-glucosidase-like [Jatropha curcas] |
| 14 |
Hb_003159_070 |
0.1759375282 |
- |
- |
Uncharacterized protein TCM_035723 [Theobroma cacao] |
| 15 |
Hb_002121_070 |
0.1791221981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005337_010 |
0.1918645731 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 17 |
Hb_005622_030 |
0.198701118 |
- |
- |
transferase, putative [Ricinus communis] |
| 18 |
Hb_000362_100 |
0.2020426224 |
- |
- |
- |
| 19 |
Hb_001089_010 |
0.2130766068 |
- |
- |
PREDICTED: GDSL esterase/lipase 5-like [Jatropha curcas] |
| 20 |
Hb_002876_100 |
0.2138568941 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |