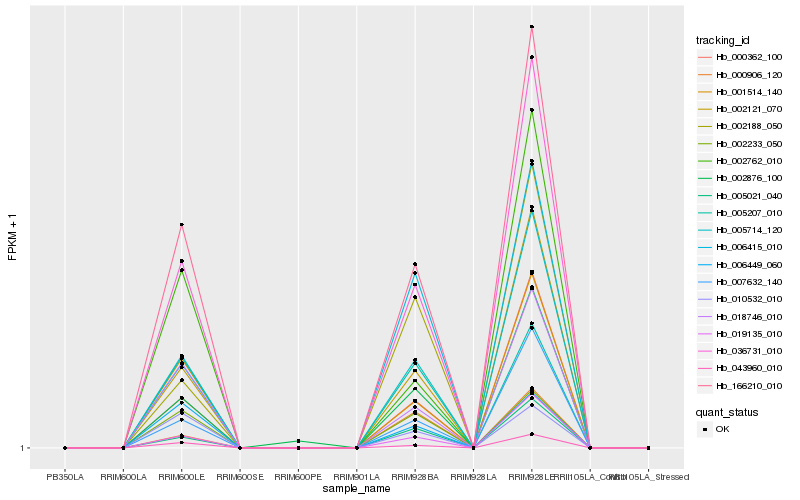
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005714_120 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_036731_010 |
0.0270027707 |
- |
- |
- |
| 3 |
Hb_002121_070 |
0.0331188234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_166210_010 |
0.0454986618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000362_100 |
0.0621932833 |
- |
- |
- |
| 6 |
Hb_001514_140 |
0.0845725089 |
- |
- |
- |
| 7 |
Hb_005021_040 |
0.0892964334 |
- |
- |
PREDICTED: uncharacterized protein LOC103848240 [Brassica rapa] |
| 8 |
Hb_007632_140 |
0.0896330576 |
- |
- |
- |
| 9 |
Hb_018746_010 |
0.0954065645 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 10 |
Hb_043960_010 |
0.0954735655 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_019135_010 |
0.1029182085 |
- |
- |
- |
| 12 |
Hb_002876_100 |
0.1137937779 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |
| 13 |
Hb_002188_050 |
0.1191091191 |
- |
- |
PREDICTED: serine carboxypeptidase-like 2 [Jatropha curcas] |
| 14 |
Hb_006415_010 |
0.1200463654 |
- |
- |
- |
| 15 |
Hb_002233_050 |
0.1251524435 |
- |
- |
PREDICTED: uncharacterized protein LOC104879391 [Vitis vinifera] |
| 16 |
Hb_002762_010 |
0.1275711949 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 17 |
Hb_005207_010 |
0.1469975213 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Jatropha curcas] |
| 18 |
Hb_000906_120 |
0.1505833497 |
- |
- |
hypothetical protein VITISV_006406 [Vitis vinifera] |
| 19 |
Hb_006449_060 |
0.1530336207 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 20 |
Hb_010532_010 |
0.1534268038 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |