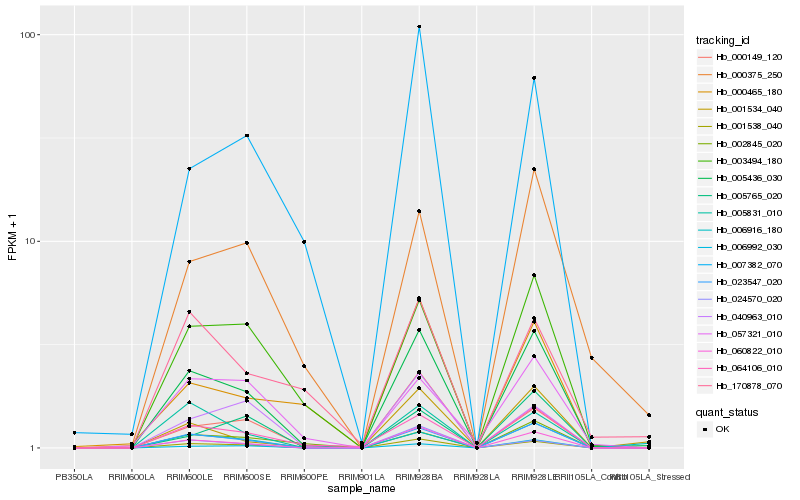
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005436_030 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74E1-like [Jatropha curcas] |
| 2 |
Hb_023547_020 |
0.0859751732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001538_040 |
0.0887796467 |
- |
- |
PREDICTED: uncharacterized protein LOC104228304 [Nicotiana sylvestris] |
| 4 |
Hb_060822_010 |
0.1002397451 |
- |
- |
- |
| 5 |
Hb_064106_010 |
0.1254657166 |
- |
- |
PREDICTED: uncharacterized protein LOC104878249 [Vitis vinifera] |
| 6 |
Hb_003494_180 |
0.1596748017 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 7 |
Hb_006992_030 |
0.1725487259 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_005831_010 |
0.1763131405 |
- |
- |
hypothetical protein JCGZ_04003 [Jatropha curcas] |
| 9 |
Hb_001534_040 |
0.1807986053 |
- |
- |
hypothetical protein JCGZ_08436 [Jatropha curcas] |
| 10 |
Hb_000149_120 |
0.1828399158 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000375_250 |
0.1872563694 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 12 |
Hb_002845_020 |
0.1884596936 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 13 |
Hb_007382_070 |
0.1917115856 |
- |
- |
PREDICTED: MAPK-interacting and spindle-stabilizing protein-like [Jatropha curcas] |
| 14 |
Hb_006916_180 |
0.192084568 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 15 |
Hb_057321_010 |
0.1962879504 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 16 |
Hb_040963_010 |
0.1964817064 |
- |
- |
PREDICTED: polyneuridine-aldehyde esterase-like [Populus euphratica] |
| 17 |
Hb_000465_180 |
0.2046815842 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005765_020 |
0.2051128613 |
- |
- |
- |
| 19 |
Hb_024570_020 |
0.2070974338 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 20 |
Hb_170878_070 |
0.2112478618 |
- |
- |
hypothetical protein EUGRSUZ_L008133, partial [Eucalyptus grandis] |