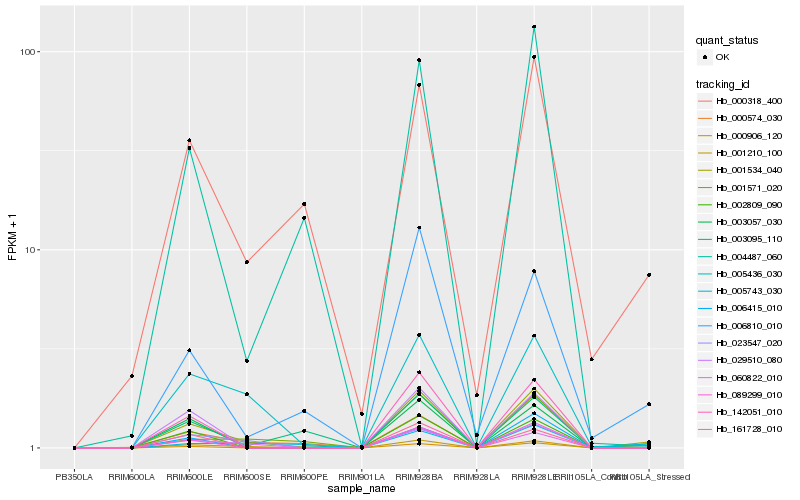
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001534_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_08436 [Jatropha curcas] |
| 2 |
Hb_060822_010 |
0.1750995117 |
- |
- |
- |
| 3 |
Hb_005436_030 |
0.1807986053 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74E1-like [Jatropha curcas] |
| 4 |
Hb_089299_010 |
0.1835229569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_142051_010 |
0.184145193 |
- |
- |
- |
| 6 |
Hb_006810_010 |
0.1860409863 |
- |
- |
hypothetical protein B456_012G018200 [Gossypium raimondii] |
| 7 |
Hb_002809_090 |
0.1948656424 |
- |
- |
- |
| 8 |
Hb_000574_030 |
0.1965540309 |
desease resistance |
Gene Name: Glyco_hydro_38 |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_029510_080 |
0.2023022152 |
- |
- |
- |
| 10 |
Hb_000318_400 |
0.2027407246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003057_030 |
0.2042587354 |
- |
- |
hypothetical protein JCGZ_06668 [Jatropha curcas] |
| 12 |
Hb_000906_120 |
0.2051745544 |
- |
- |
hypothetical protein VITISV_006406 [Vitis vinifera] |
| 13 |
Hb_004487_060 |
0.205762233 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_006415_010 |
0.2065915027 |
- |
- |
- |
| 15 |
Hb_001571_020 |
0.2076501329 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850-like [Citrus sinensis] |
| 16 |
Hb_005743_030 |
0.2083919887 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 17 |
Hb_023547_020 |
0.208756515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001210_100 |
0.2090551716 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 19 |
Hb_003095_110 |
0.2103468059 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 20 |
Hb_161728_010 |
0.2138399736 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |