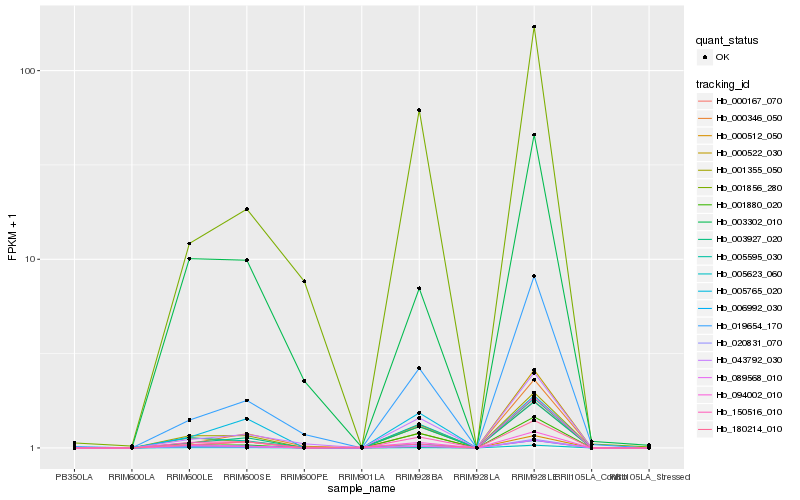
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001355_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_005595_030 |
0.0757495765 |
- |
- |
hypothetical protein EUGRSUZ_A00624 [Eucalyptus grandis] |
| 3 |
Hb_003927_020 |
0.0913705305 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 4 |
Hb_006992_030 |
0.0919477854 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_150516_010 |
0.1266087302 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_001880_020 |
0.1361798255 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 7 |
Hb_043792_030 |
0.141826526 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 8 |
Hb_094002_010 |
0.1433379344 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 9 |
Hb_000346_050 |
0.1488480111 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000167_070 |
0.1606536114 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 11 |
Hb_019654_170 |
0.160852682 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 12 |
Hb_003302_010 |
0.1622259882 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 13 |
Hb_001856_280 |
0.1626588831 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 14 |
Hb_089568_010 |
0.1643897809 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 15 |
Hb_020831_070 |
0.1681783188 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 16 |
Hb_005623_060 |
0.1686492038 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_180214_010 |
0.1757127398 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 18 |
Hb_005765_020 |
0.1758739637 |
- |
- |
- |
| 19 |
Hb_000512_050 |
0.1794701895 |
- |
- |
hypothetical protein VITISV_005640 [Vitis vinifera] |
| 20 |
Hb_000522_030 |
0.1888197287 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |