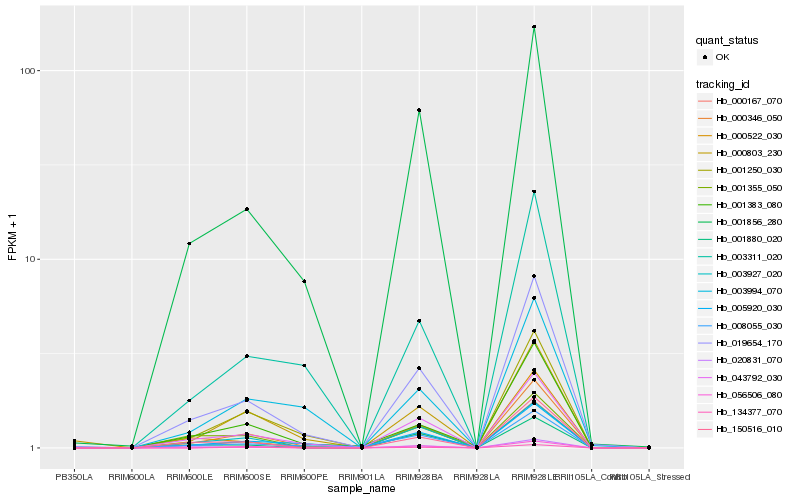
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000346_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_043792_030 |
0.0680997428 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 3 |
Hb_019654_170 |
0.0728472956 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 4 |
Hb_001856_280 |
0.0958860209 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 5 |
Hb_003927_020 |
0.1157467763 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 6 |
Hb_150516_010 |
0.1243752089 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_000167_070 |
0.1294076044 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 8 |
Hb_000803_230 |
0.1302620561 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 9 |
Hb_005920_030 |
0.1322633294 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 10 |
Hb_001383_080 |
0.133166388 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 11 |
Hb_003311_020 |
0.1344812016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001880_020 |
0.1466744564 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 13 |
Hb_001355_050 |
0.1488480111 |
- |
- |
- |
| 14 |
Hb_020831_070 |
0.1504289528 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 15 |
Hb_008055_030 |
0.1526392443 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 16 |
Hb_001250_030 |
0.1529890253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_056506_080 |
0.1542761565 |
- |
- |
PREDICTED: uncharacterized protein LOC104888944 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_003994_070 |
0.1601812909 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 19 |
Hb_000522_030 |
0.161086647 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 20 |
Hb_134377_070 |
0.1627122091 |
- |
- |
- |