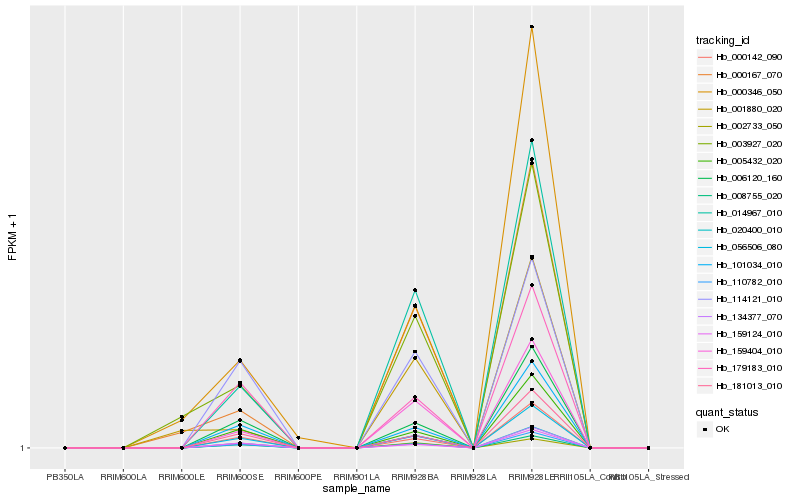
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014967_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_159404_010 |
0.0157170963 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 3 |
Hb_020400_010 |
0.060119832 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |
| 4 |
Hb_008755_020 |
0.0835738762 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_056506_080 |
0.0977750828 |
- |
- |
PREDICTED: uncharacterized protein LOC104888944 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_110782_010 |
0.105447461 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 7 |
Hb_000167_070 |
0.1245986394 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 8 |
Hb_006120_160 |
0.1426927452 |
- |
- |
- |
| 9 |
Hb_114121_010 |
0.1439453829 |
- |
- |
- |
| 10 |
Hb_101034_010 |
0.145031898 |
- |
- |
- |
| 11 |
Hb_002733_050 |
0.1471446351 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_005432_020 |
0.1472229782 |
- |
- |
- |
| 13 |
Hb_181013_010 |
0.149733183 |
- |
- |
PREDICTED: uncharacterized protein LOC105629028 [Jatropha curcas] |
| 14 |
Hb_000142_090 |
0.1518885463 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 15 |
Hb_134377_070 |
0.1559046056 |
- |
- |
- |
| 16 |
Hb_003927_020 |
0.1630986282 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 17 |
Hb_179183_010 |
0.1637923765 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 18 |
Hb_159124_010 |
0.1709183565 |
- |
- |
PREDICTED: uncharacterized protein LOC103332224 [Prunus mume] |
| 19 |
Hb_001880_020 |
0.1722875917 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 20 |
Hb_000346_050 |
0.1726028102 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |