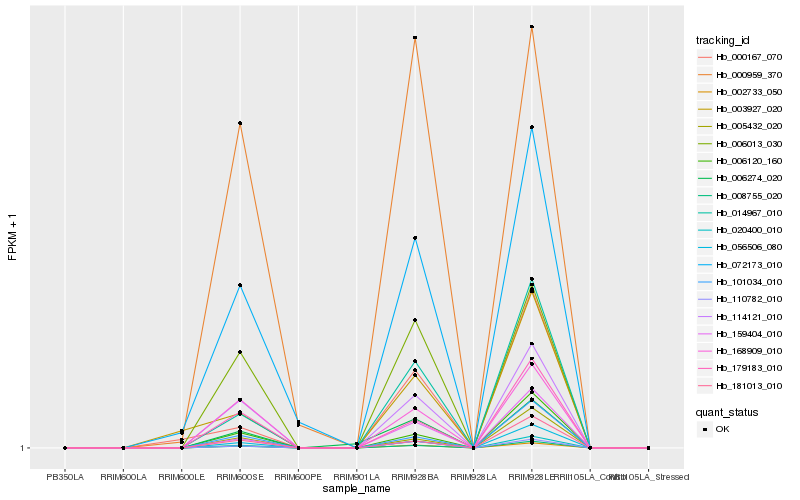
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020400_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |
| 2 |
Hb_110782_010 |
0.0562865611 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 3 |
Hb_014967_010 |
0.060119832 |
- |
- |
- |
| 4 |
Hb_159404_010 |
0.0757705385 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 5 |
Hb_008755_020 |
0.0891009964 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002733_050 |
0.1070832768 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 7 |
Hb_114121_010 |
0.1165281618 |
- |
- |
- |
| 8 |
Hb_056506_080 |
0.1262545544 |
- |
- |
PREDICTED: uncharacterized protein LOC104888944 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_006013_030 |
0.1287977238 |
- |
- |
- |
| 10 |
Hb_000167_070 |
0.1520766908 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 11 |
Hb_006274_020 |
0.1532934435 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 12 |
Hb_072173_010 |
0.1595188472 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 13 |
Hb_179183_010 |
0.1659920089 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 14 |
Hb_006120_160 |
0.1663530308 |
- |
- |
- |
| 15 |
Hb_168909_010 |
0.1693724253 |
- |
- |
PREDICTED: uncharacterized protein LOC105640156 [Jatropha curcas] |
| 16 |
Hb_101034_010 |
0.1707807641 |
- |
- |
- |
| 17 |
Hb_005432_020 |
0.1746565945 |
- |
- |
- |
| 18 |
Hb_003927_020 |
0.1748558895 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 19 |
Hb_181013_010 |
0.178859813 |
- |
- |
PREDICTED: uncharacterized protein LOC105629028 [Jatropha curcas] |
| 20 |
Hb_000959_370 |
0.1807177329 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |