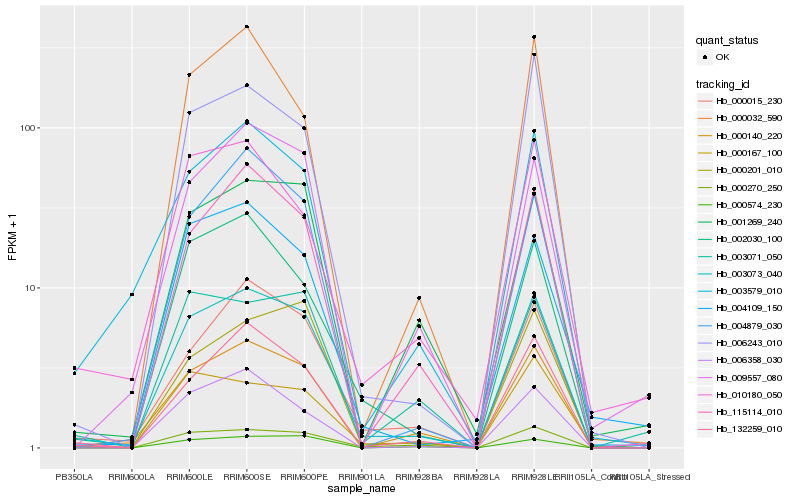
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003073_040 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 2 |
Hb_000140_220 |
0.0955120496 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 3 |
Hb_132259_010 |
0.10748746 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 4 |
Hb_000015_230 |
0.1141324976 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 5 |
Hb_115114_010 |
0.1191415802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000270_250 |
0.1196488754 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s07900g [Populus trichocarpa] |
| 7 |
Hb_002030_100 |
0.1244010928 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 8 |
Hb_003579_010 |
0.1262174627 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 9 |
Hb_001269_240 |
0.1328749811 |
- |
- |
aquaporin NIP6.1 family protein [Populus trichocarpa] |
| 10 |
Hb_009557_080 |
0.1330232986 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 11 |
Hb_006358_030 |
0.1347501342 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 12 |
Hb_004879_030 |
0.1354308822 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 13 |
Hb_000167_100 |
0.1367850815 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_006243_010 |
0.1385652391 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |
| 15 |
Hb_004109_150 |
0.1404987925 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 16 |
Hb_000574_230 |
0.1452716573 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EXS-like isoform X2 [Vitis vinifera] |
| 17 |
Hb_000201_010 |
0.1473898965 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000032_590 |
0.1498583032 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 19 |
Hb_010180_050 |
0.1526162552 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 20 |
Hb_003071_050 |
0.1536343474 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |