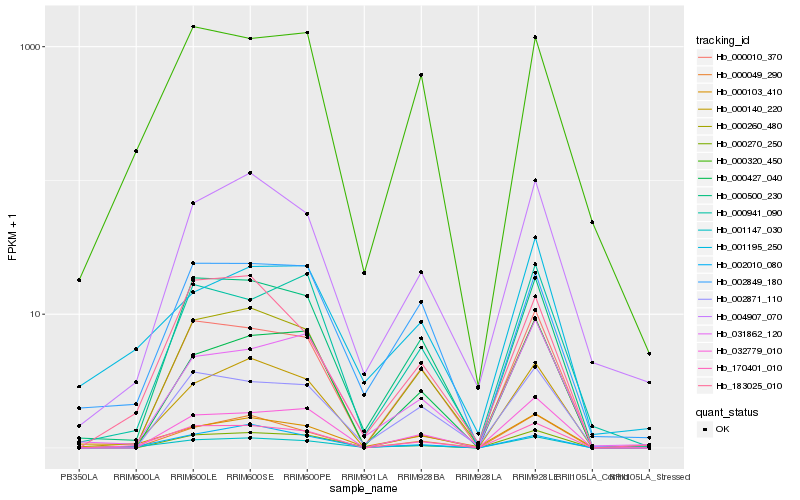
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001147_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 2 |
Hb_170401_010 |
0.0859226556 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 3 |
Hb_000500_230 |
0.1239460875 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 4 |
Hb_000270_250 |
0.1248095817 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s07900g [Populus trichocarpa] |
| 5 |
Hb_004907_070 |
0.1375366116 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 6 |
Hb_002010_080 |
0.1376797781 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 7 |
Hb_000010_370 |
0.1410838908 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 8 |
Hb_000260_480 |
0.1416245706 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 9 |
Hb_000140_220 |
0.1448987545 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 10 |
Hb_000049_290 |
0.146694042 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_000103_410 |
0.1495417544 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 12 |
Hb_183025_010 |
0.1506535303 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 13 |
Hb_000320_450 |
0.1537252442 |
- |
- |
secretory peroxidase [Hevea brasiliensis] |
| 14 |
Hb_000427_040 |
0.1550455743 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 15 |
Hb_002871_110 |
0.1550783635 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 16 |
Hb_002849_180 |
0.1564687036 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 17 |
Hb_031862_120 |
0.1566600884 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 18 |
Hb_000941_090 |
0.1584482933 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 19 |
Hb_032779_010 |
0.1586904075 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001195_250 |
0.1607410369 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |