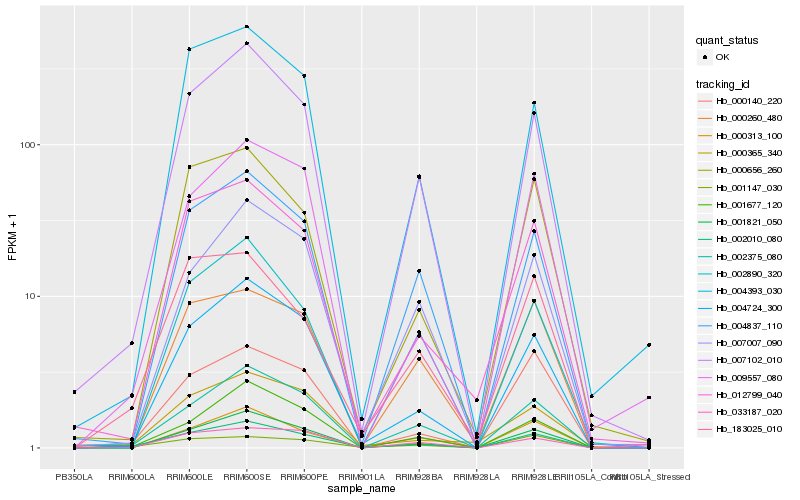
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002010_080 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 2 |
Hb_007102_010 |
0.1036368887 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 3 |
Hb_004724_300 |
0.1117281068 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 4 |
Hb_001821_050 |
0.1189362322 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 5 |
Hb_002375_080 |
0.1224523969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004837_110 |
0.1300642181 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_012799_040 |
0.134841235 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 8 |
Hb_183025_010 |
0.1350505538 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 9 |
Hb_001677_120 |
0.1372958908 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_001147_030 |
0.1376797781 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 11 |
Hb_000656_260 |
0.1412466495 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 12 |
Hb_033187_020 |
0.1420910911 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 13 |
Hb_000140_220 |
0.1426423634 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 14 |
Hb_009557_080 |
0.1436426712 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 15 |
Hb_004393_030 |
0.1447848274 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 16 |
Hb_007007_090 |
0.1448021497 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 17 |
Hb_000365_340 |
0.1465322275 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 18 |
Hb_000260_480 |
0.1486877672 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 19 |
Hb_002890_320 |
0.1494794834 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 20 |
Hb_000313_100 |
0.1496471487 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |