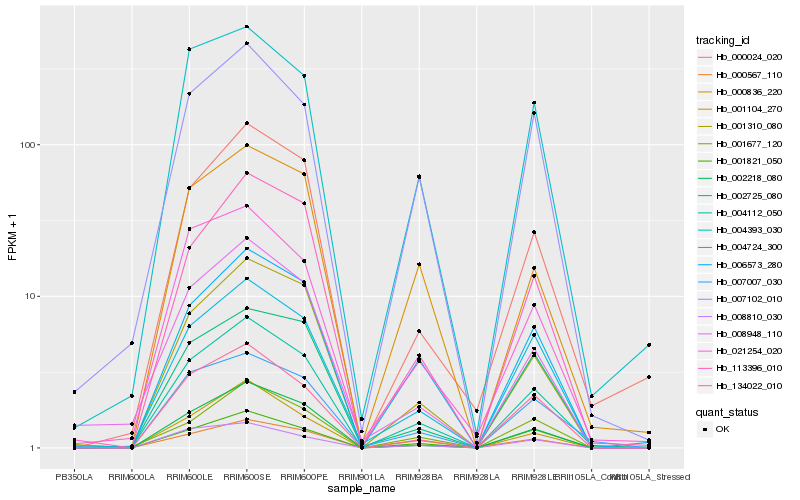
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004112_050 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 2 |
Hb_001310_080 |
0.0834889188 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 3 |
Hb_004724_300 |
0.093440604 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 4 |
Hb_002218_080 |
0.0936725328 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 5 |
Hb_007102_010 |
0.0944057952 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 6 |
Hb_007007_030 |
0.0957339348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_113396_010 |
0.0963027142 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_006573_280 |
0.0967404364 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 9 |
Hb_001104_270 |
0.0968828589 |
- |
- |
hypothetical protein POPTR_0007s13980g [Populus trichocarpa] |
| 10 |
Hb_008948_110 |
0.1008370786 |
- |
- |
PREDICTED: peroxidase 25 [Jatropha curcas] |
| 11 |
Hb_134022_010 |
0.1014157301 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 12 |
Hb_000024_020 |
0.1022021406 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 13 |
Hb_004393_030 |
0.1032279241 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 14 |
Hb_021254_020 |
0.1085681569 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_001821_050 |
0.1109012138 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 16 |
Hb_000836_220 |
0.1112987189 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001677_120 |
0.1127658551 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 18 |
Hb_000567_110 |
0.1131998941 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 19 |
Hb_008810_030 |
0.1185101288 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_002725_080 |
0.1196458738 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |