| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
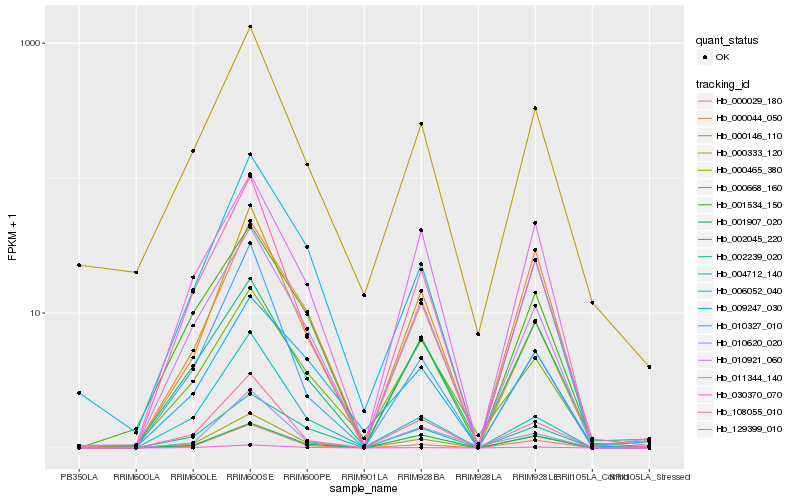
Hb_000465_380 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000029_180 |
0.077006035 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 3 |
Hb_108055_010 |
0.0844859477 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_010921_060 |
0.0943185155 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_030370_070 |
0.0955731752 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_129399_010 |
0.1026260848 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_002239_020 |
0.1069713462 |
- |
- |
MLO, putative [Theobroma cacao] |
| 8 |
Hb_011344_140 |
0.1074983936 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 9 |
Hb_000146_110 |
0.126449181 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 10 |
Hb_002045_220 |
0.1301089572 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000333_120 |
0.1340791096 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 12 |
Hb_001907_020 |
0.1341982715 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 13 |
Hb_000044_050 |
0.1355093941 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001534_150 |
0.1395240927 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 15 |
Hb_010327_010 |
0.1413717795 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_006052_040 |
0.143546595 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 17 |
Hb_009247_030 |
0.1439791053 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 18 |
Hb_000668_160 |
0.1447418531 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 19 |
Hb_004712_140 |
0.1474043747 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 20 |
Hb_010620_020 |
0.1488501136 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |