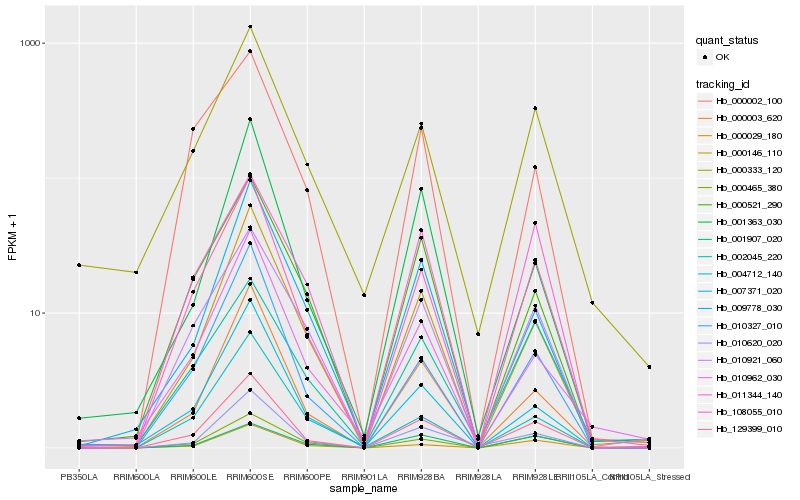
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129399_010 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_108055_010 |
0.0735569773 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000465_380 |
0.1026260848 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_010620_020 |
0.1079113483 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000146_110 |
0.1093708968 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 6 |
Hb_000003_620 |
0.1153594069 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 7 |
Hb_010921_060 |
0.1182769083 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_010327_010 |
0.1274738975 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_000029_180 |
0.1354638057 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 10 |
Hb_010962_030 |
0.1433498379 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_000521_290 |
0.1435315066 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 12 |
Hb_007371_020 |
0.1439953175 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_011344_140 |
0.1459245607 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 14 |
Hb_000333_120 |
0.1473192176 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 15 |
Hb_009778_030 |
0.1477753968 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 16 |
Hb_002045_220 |
0.1505253239 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004712_140 |
0.1506932106 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 18 |
Hb_001907_020 |
0.1548188329 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 19 |
Hb_000002_100 |
0.1579564877 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 20 |
Hb_001363_030 |
0.1611790126 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |