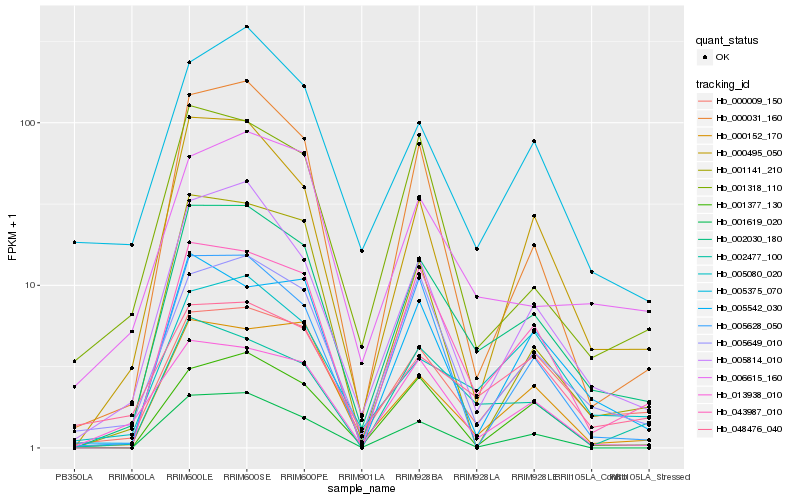
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002030_180 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_043987_010 |
0.115306364 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 3 |
Hb_005814_010 |
0.1193882053 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000495_050 |
0.1272800482 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 5 |
Hb_000031_160 |
0.1338571343 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 6 |
Hb_002477_100 |
0.1386033304 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 7 |
Hb_000009_150 |
0.1392239224 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 8 |
Hb_005649_010 |
0.1400902482 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 9 |
Hb_000152_170 |
0.1469454441 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 10 |
Hb_005080_020 |
0.1510003412 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 11 |
Hb_006615_160 |
0.151157865 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 12 |
Hb_001141_210 |
0.1514541348 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 13 |
Hb_001619_020 |
0.152028461 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 14 |
Hb_013938_010 |
0.1543556891 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 15 |
Hb_001318_110 |
0.1555946395 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 16 |
Hb_048476_040 |
0.1568606842 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 17 |
Hb_001377_130 |
0.1583714112 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 18 |
Hb_005628_050 |
0.1599199574 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_005375_070 |
0.1615628943 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 20 |
Hb_005542_030 |
0.1627855909 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 isoform X2 [Jatropha curcas] |