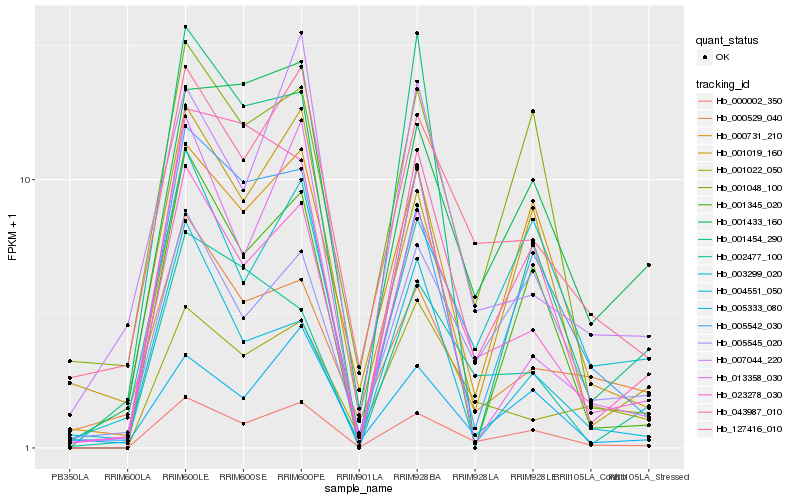
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023278_030 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000002_350 |
0.0885206301 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 3 |
Hb_127416_010 |
0.110012401 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 4 |
Hb_001022_050 |
0.1544063829 |
- |
- |
hypothetical protein POPTR_0016s07780g [Populus trichocarpa] |
| 5 |
Hb_005545_020 |
0.1551805754 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 6 |
Hb_002477_100 |
0.1601784708 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 7 |
Hb_000529_040 |
0.1608993455 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 8 |
Hb_000731_210 |
0.1667309526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003299_020 |
0.1669606465 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 10 |
Hb_005542_030 |
0.1681785709 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 isoform X2 [Jatropha curcas] |
| 11 |
Hb_043987_010 |
0.1685508499 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 12 |
Hb_007044_220 |
0.172025475 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_001048_100 |
0.1751105279 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 14 |
Hb_013358_030 |
0.1765362875 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 15 |
Hb_005333_080 |
0.1766474489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004551_050 |
0.1777600075 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001019_160 |
0.1778006863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001454_290 |
0.1781058013 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 19 |
Hb_001433_160 |
0.1791596603 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 20 |
Hb_001345_020 |
0.1794939847 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |