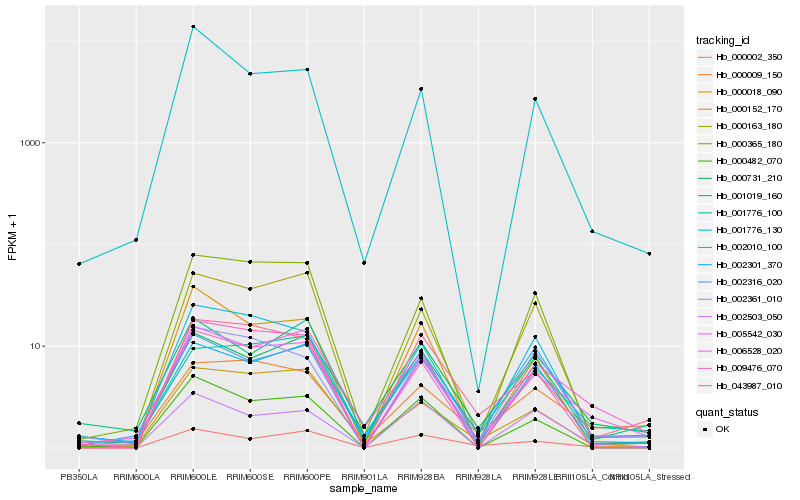
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005542_030 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 isoform X2 [Jatropha curcas] |
| 2 |
Hb_006528_020 |
0.1201540737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_043987_010 |
0.1353802686 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 4 |
Hb_000163_180 |
0.1359621736 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 5 |
Hb_000152_170 |
0.1381746145 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 6 |
Hb_002361_010 |
0.1396924008 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000482_070 |
0.1405388268 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 8 |
Hb_000002_350 |
0.1414302125 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 9 |
Hb_002010_100 |
0.144435134 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 10 |
Hb_000018_090 |
0.1448091697 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001776_100 |
0.1457195291 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000365_180 |
0.1477306386 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 13 |
Hb_001776_130 |
0.1479256592 |
- |
- |
metallothionein [Hevea brasiliensis] |
| 14 |
Hb_009476_070 |
0.1480901476 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 15 |
Hb_000731_210 |
0.1490273968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002301_370 |
0.1507182656 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 17 |
Hb_000009_150 |
0.1518245453 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 18 |
Hb_002316_020 |
0.153572244 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 19 |
Hb_002503_050 |
0.154986521 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 20 |
Hb_001019_160 |
0.1564883764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |