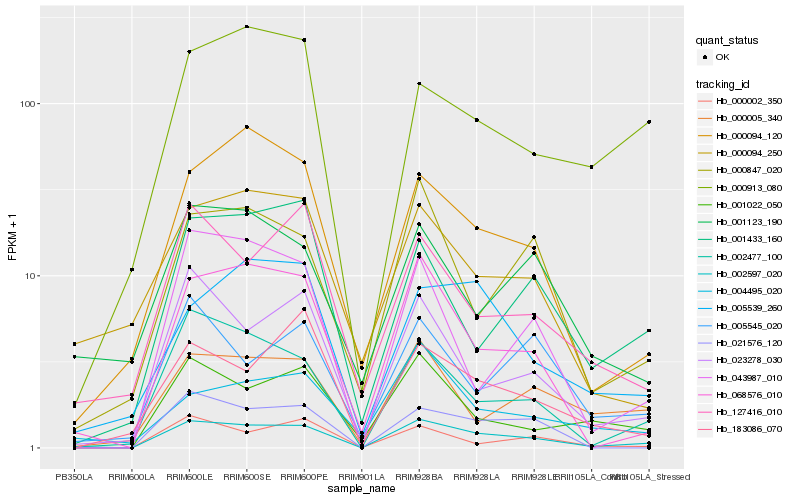
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002597_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 2 |
Hb_068576_010 |
0.16099826 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 3 |
Hb_002477_100 |
0.1638579864 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 4 |
Hb_023278_030 |
0.1865558135 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_021576_120 |
0.1874318061 |
- |
- |
DNA-directed RNA polymerase subunit, putative [Ricinus communis] |
| 6 |
Hb_000005_340 |
0.1874558813 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000847_020 |
0.1893996152 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 8 |
Hb_000094_120 |
0.1932263006 |
- |
- |
PREDICTED: uncharacterized protein At1g01500 [Jatropha curcas] |
| 9 |
Hb_001022_050 |
0.1933137041 |
- |
- |
hypothetical protein POPTR_0016s07780g [Populus trichocarpa] |
| 10 |
Hb_005539_260 |
0.1969477283 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |
| 11 |
Hb_004495_020 |
0.2010615375 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_127416_010 |
0.2015758643 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_043987_010 |
0.2039265078 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 14 |
Hb_001433_160 |
0.2047464223 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 15 |
Hb_000002_350 |
0.2061824239 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 16 |
Hb_000094_250 |
0.2063037126 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 17 |
Hb_001123_190 |
0.2078310353 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 18 |
Hb_000913_080 |
0.2092265564 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 19 |
Hb_183086_070 |
0.2097256585 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like [Jatropha curcas] |
| 20 |
Hb_005545_020 |
0.2099269859 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |