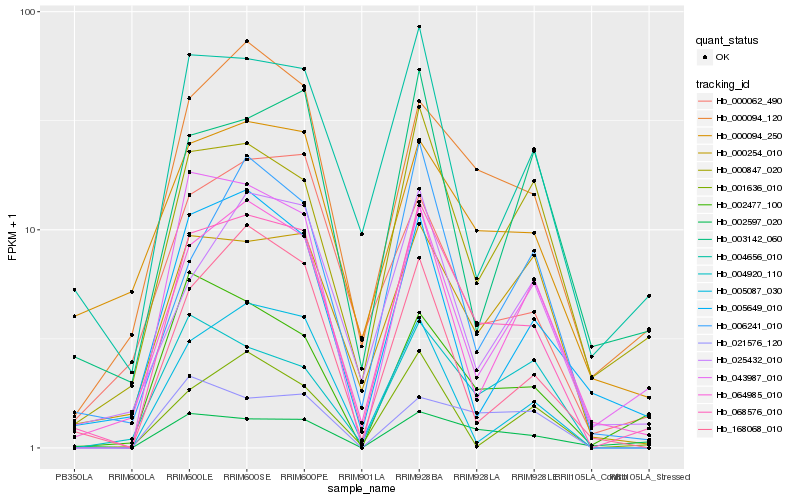
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068576_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 2 |
Hb_000094_120 |
0.1533893212 |
- |
- |
PREDICTED: uncharacterized protein At1g01500 [Jatropha curcas] |
| 3 |
Hb_004656_010 |
0.1603502056 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 4 |
Hb_002597_020 |
0.16099826 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 5 |
Hb_064985_010 |
0.161195892 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 6 |
Hb_168068_010 |
0.168725388 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 7 |
Hb_000847_020 |
0.1722026442 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 8 |
Hb_043987_010 |
0.1727904181 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 9 |
Hb_000254_010 |
0.1744242207 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 10 |
Hb_005087_030 |
0.1744436409 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 11 |
Hb_025432_010 |
0.1788571108 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 12 |
Hb_000094_250 |
0.1789096795 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 13 |
Hb_006241_010 |
0.1805064857 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 14 |
Hb_000062_490 |
0.1828822419 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002477_100 |
0.1829590314 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 16 |
Hb_005649_010 |
0.1843538774 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 17 |
Hb_001636_010 |
0.1911771571 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 18 |
Hb_003142_060 |
0.1912990126 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_004920_110 |
0.1924005844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_021576_120 |
0.192535753 |
- |
- |
DNA-directed RNA polymerase subunit, putative [Ricinus communis] |