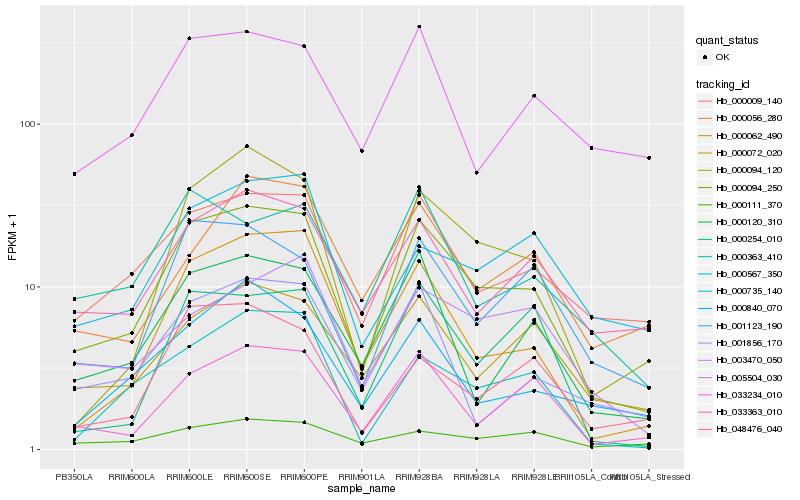
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000009_140 |
0.1348697419 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 3 |
Hb_033234_010 |
0.1373540392 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 4 |
Hb_000062_490 |
0.137505701 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000094_120 |
0.1396195428 |
- |
- |
PREDICTED: uncharacterized protein At1g01500 [Jatropha curcas] |
| 6 |
Hb_000120_310 |
0.1412999811 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 7 |
Hb_001123_190 |
0.1432140502 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 8 |
Hb_033363_010 |
0.1459351173 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 9 |
Hb_000072_020 |
0.1497230227 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000567_350 |
0.1535145063 |
- |
- |
PREDICTED: probable acyl-activating enzyme 6 [Jatropha curcas] |
| 11 |
Hb_000363_410 |
0.1582221922 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 12 |
Hb_000111_370 |
0.1586373874 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 13 |
Hb_000254_010 |
0.1613378909 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 14 |
Hb_048476_040 |
0.1617774383 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 15 |
Hb_001856_170 |
0.162099852 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 16 |
Hb_000840_070 |
0.1621369803 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_003470_050 |
0.1641485662 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000735_140 |
0.167150264 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 19 |
Hb_000056_280 |
0.1691310019 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 20 |
Hb_005504_030 |
0.1696089778 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |