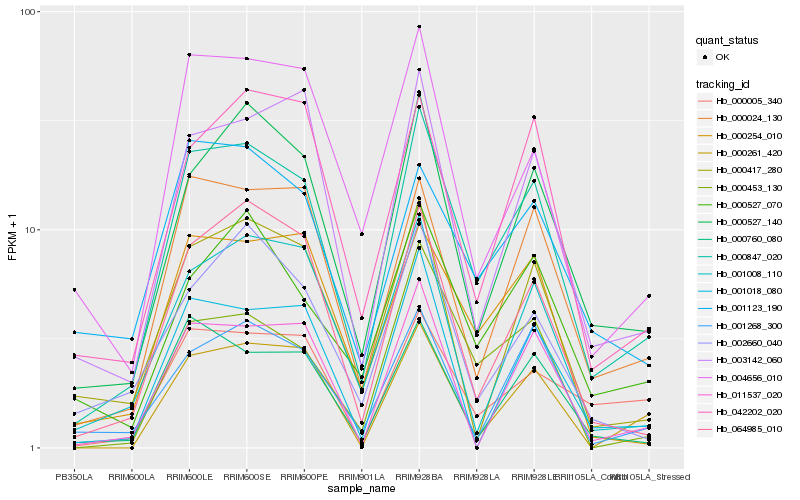
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000847_020 |
0.0 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 2 |
Hb_000527_140 |
0.1155096628 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 3 |
Hb_000024_130 |
0.1247254797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_064985_010 |
0.1329201544 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 5 |
Hb_003142_060 |
0.1392780883 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000527_070 |
0.1428980445 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 7 |
Hb_004656_010 |
0.1436223348 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 8 |
Hb_000254_010 |
0.143688323 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 9 |
Hb_042202_020 |
0.1448786824 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 10 |
Hb_000760_080 |
0.1453535757 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 11 |
Hb_000005_340 |
0.1464851806 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001008_110 |
0.1504073651 |
- |
- |
- |
| 13 |
Hb_000453_130 |
0.15236244 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_001268_300 |
0.1531853491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000417_280 |
0.1569531531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001018_080 |
0.1574122634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002660_040 |
0.1579108513 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001123_190 |
0.158513229 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 19 |
Hb_011537_020 |
0.1588650913 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 20 |
Hb_000261_420 |
0.1599834602 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |