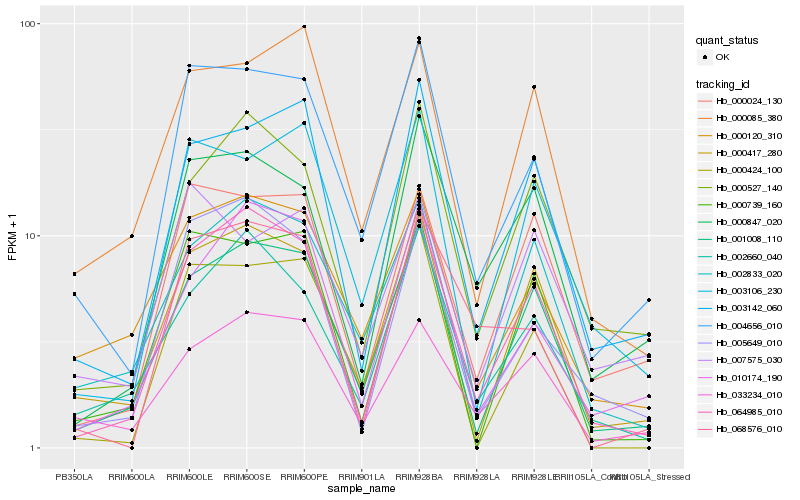
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004656_010 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 2 |
Hb_000120_310 |
0.1272772572 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 3 |
Hb_003106_230 |
0.1277133015 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 4 |
Hb_001008_110 |
0.129114445 |
- |
- |
- |
| 5 |
Hb_000417_280 |
0.1301664362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003142_060 |
0.1331324553 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_064985_010 |
0.1410052953 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 8 |
Hb_000847_020 |
0.1436223348 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 9 |
Hb_000024_130 |
0.1445854298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_033234_010 |
0.1458100054 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 11 |
Hb_000424_100 |
0.1477069835 |
desease resistance |
Gene Name: G-alpha |
GTP-binding protein (p) alpha subunit, gpa1, putative [Ricinus communis] |
| 12 |
Hb_005649_010 |
0.1516126503 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 13 |
Hb_000527_140 |
0.1574881359 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 14 |
Hb_000085_380 |
0.1575241254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007575_030 |
0.1577891748 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 16 |
Hb_010174_190 |
0.158311465 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 17 |
Hb_000739_160 |
0.1585130743 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_002833_020 |
0.1589096395 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_002660_040 |
0.1603330318 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_068576_010 |
0.1603502056 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |