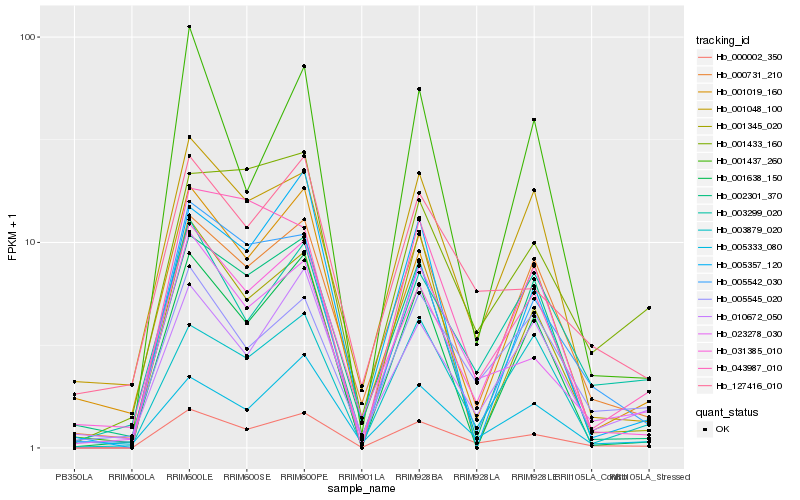
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_350 |
0.0 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 2 |
Hb_023278_030 |
0.0885206301 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000731_210 |
0.1246897994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_127416_010 |
0.1368864951 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 5 |
Hb_005542_030 |
0.1414302125 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003299_020 |
0.1462018596 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 7 |
Hb_005545_020 |
0.1483505152 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 8 |
Hb_005333_080 |
0.1501181621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010672_050 |
0.1508023482 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001019_160 |
0.1534455367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003879_020 |
0.1539220413 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 12 |
Hb_001433_160 |
0.1573638409 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 13 |
Hb_001048_100 |
0.1577381849 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 14 |
Hb_001437_260 |
0.1583029901 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 15 |
Hb_043987_010 |
0.1594087886 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 16 |
Hb_031385_010 |
0.1604017187 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 17 |
Hb_001345_020 |
0.1610037733 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 18 |
Hb_001638_150 |
0.1629642594 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 19 |
Hb_002301_370 |
0.1650294706 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 20 |
Hb_005357_120 |
0.1678413728 |
transcription factor |
TF Family: MYB-related |
- |