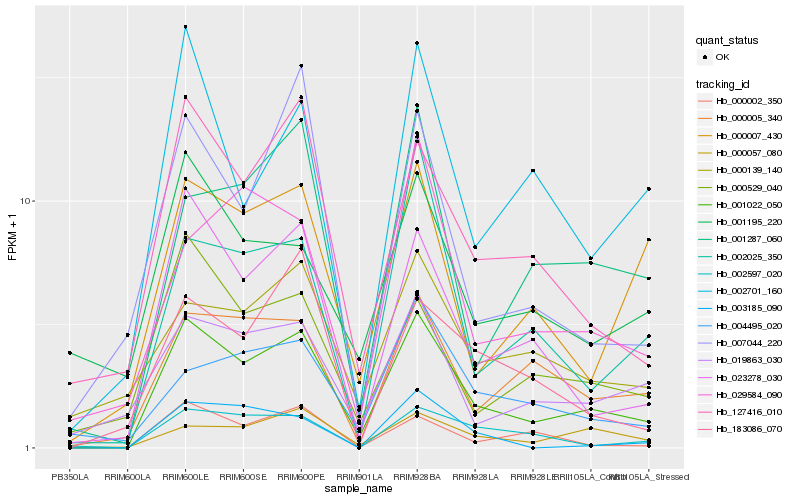
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001022_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s07780g [Populus trichocarpa] |
| 2 |
Hb_023278_030 |
0.1544063829 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_127416_010 |
0.1677993547 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 4 |
Hb_000005_340 |
0.1767461221 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000057_080 |
0.1829441687 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 6 |
Hb_000002_350 |
0.1850095006 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 7 |
Hb_019863_030 |
0.1892138083 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 8 |
Hb_001287_060 |
0.1898688488 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 9 |
Hb_000529_040 |
0.19285761 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 10 |
Hb_002597_020 |
0.1933137041 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_002701_160 |
0.1946634525 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 12 |
Hb_000007_430 |
0.1982187547 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 13 |
Hb_004495_020 |
0.1997496949 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_183086_070 |
0.202344869 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like [Jatropha curcas] |
| 15 |
Hb_002025_350 |
0.2024518109 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 16 |
Hb_007044_220 |
0.2041050381 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000139_140 |
0.2073566801 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 18 |
Hb_003185_090 |
0.2079504582 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 19 |
Hb_001195_220 |
0.2095258149 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 20 |
Hb_029584_090 |
0.209692521 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |