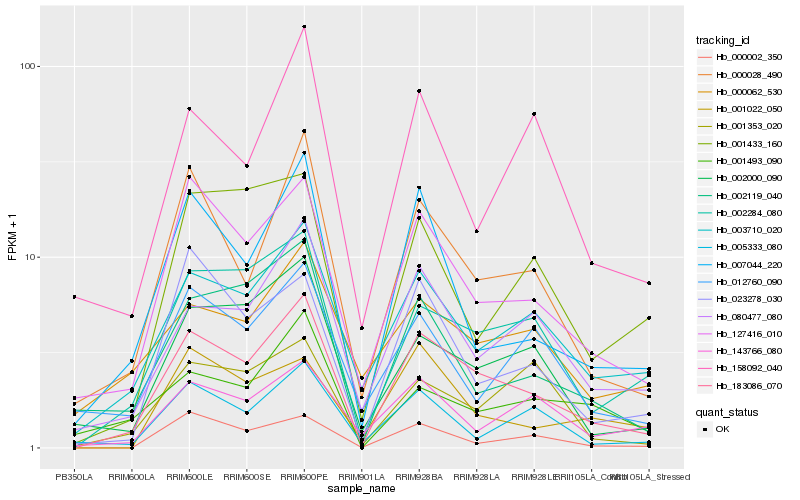
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183086_070 |
0.0 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like [Jatropha curcas] |
| 2 |
Hb_127416_010 |
0.1358821146 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 3 |
Hb_000028_490 |
0.1432074654 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 4 |
Hb_002284_080 |
0.1535101247 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_003710_020 |
0.1558273708 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007044_220 |
0.15937298 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_080477_080 |
0.1647832056 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 8 |
Hb_002000_090 |
0.1737153011 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 9 |
Hb_002119_040 |
0.1805255146 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000002_350 |
0.1816610003 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 11 |
Hb_023278_030 |
0.1831463139 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_005333_080 |
0.1926751169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_158092_040 |
0.1961868649 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 14 |
Hb_001353_020 |
0.200722775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000062_530 |
0.2021806982 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 16 |
Hb_001022_050 |
0.202344869 |
- |
- |
hypothetical protein POPTR_0016s07780g [Populus trichocarpa] |
| 17 |
Hb_012760_090 |
0.2027559205 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 18 |
Hb_143766_080 |
0.2030044769 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001433_160 |
0.2033525083 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 20 |
Hb_001493_090 |
0.2036900753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |