| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
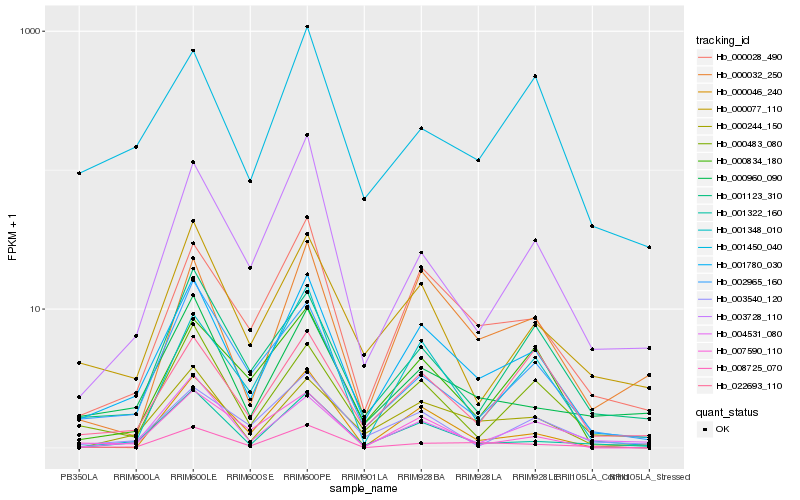
Hb_001780_030 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 2 |
Hb_000028_490 |
0.1198191646 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 3 |
Hb_000244_150 |
0.1294658122 |
- |
- |
PREDICTED: chromatin assembly factor 1 subunit FAS2 [Jatropha curcas] |
| 4 |
Hb_000483_080 |
0.1546384574 |
- |
- |
hypothetical protein CISIN_1g000833mg [Citrus sinensis] |
| 5 |
Hb_001322_160 |
0.1563577766 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |
| 6 |
Hb_008725_070 |
0.1563919402 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 7 |
Hb_000077_110 |
0.1590611385 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |
| 8 |
Hb_000960_090 |
0.1631513186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000032_250 |
0.1655811976 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 10 |
Hb_002965_160 |
0.1662685308 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 11 |
Hb_001123_310 |
0.1685605348 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 12 |
Hb_007590_110 |
0.1704616335 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 13 |
Hb_022693_110 |
0.1710059521 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 14 |
Hb_000834_180 |
0.1710452206 |
- |
- |
PREDICTED: kinesin-13A-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001450_040 |
0.1720263691 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 16 |
Hb_001348_010 |
0.1735361039 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 17 |
Hb_000046_240 |
0.1748243688 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 18 |
Hb_003728_110 |
0.1750151673 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 19 |
Hb_003540_120 |
0.1754343274 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_004531_080 |
0.1754444161 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |