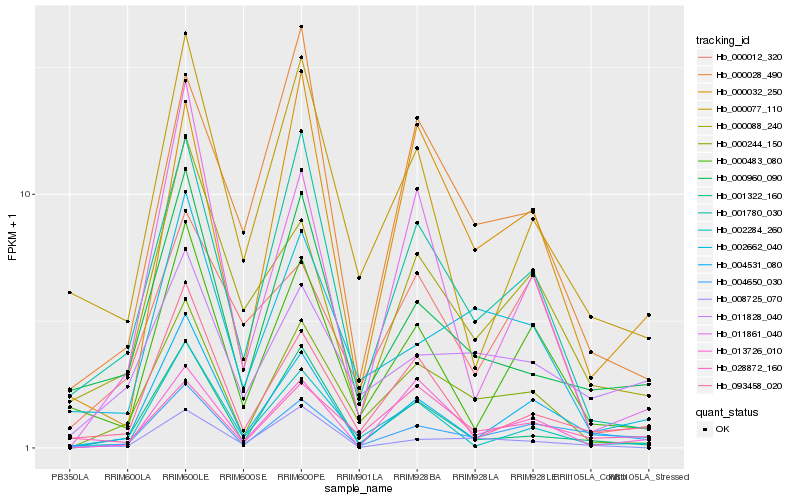
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000244_150 |
0.0 |
- |
- |
PREDICTED: chromatin assembly factor 1 subunit FAS2 [Jatropha curcas] |
| 2 |
Hb_001780_030 |
0.1294658122 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 3 |
Hb_002662_040 |
0.1656022358 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 4 |
Hb_001322_160 |
0.1876126667 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |
| 5 |
Hb_013726_010 |
0.1913570998 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000028_490 |
0.1918025904 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 7 |
Hb_004531_080 |
0.1925362491 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 8 |
Hb_000960_090 |
0.1963416583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000032_250 |
0.1972911442 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 10 |
Hb_002284_260 |
0.1991486339 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 11 |
Hb_011861_040 |
0.2001022682 |
- |
- |
PREDICTED: pheromone-processing carboxypeptidase KEX1-like [Jatropha curcas] |
| 12 |
Hb_000088_240 |
0.2029506666 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000483_080 |
0.2037564449 |
- |
- |
hypothetical protein CISIN_1g000833mg [Citrus sinensis] |
| 14 |
Hb_093458_020 |
0.2043700166 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 15 |
Hb_004650_030 |
0.20491137 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011828_040 |
0.2058992763 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 17 |
Hb_008725_070 |
0.2074607015 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 18 |
Hb_000012_320 |
0.2086660231 |
- |
- |
PREDICTED: uncharacterized protein LOC105638164 [Jatropha curcas] |
| 19 |
Hb_028872_160 |
0.2088848348 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |
| 20 |
Hb_000077_110 |
0.2093502336 |
- |
- |
PREDICTED: uncharacterized protein LOC105646935 [Jatropha curcas] |