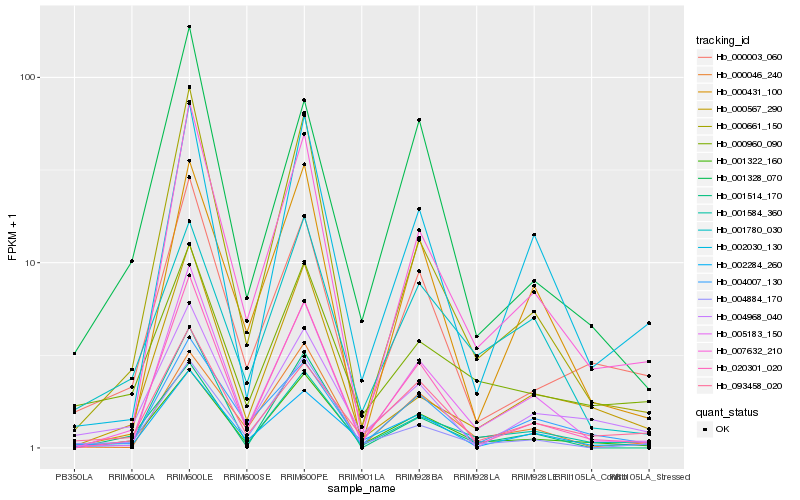
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001322_160 |
0.0 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |
| 2 |
Hb_020301_020 |
0.1232206058 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 3 |
Hb_000661_150 |
0.1368630251 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 4 |
Hb_000003_060 |
0.1401883039 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 5 |
Hb_002284_260 |
0.1418382533 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 6 |
Hb_007632_210 |
0.1456835674 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 7 |
Hb_005183_150 |
0.1469264523 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 8 |
Hb_000960_090 |
0.1519247771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002030_130 |
0.1534118619 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 10 |
Hb_000046_240 |
0.1548758021 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 11 |
Hb_093458_020 |
0.155403882 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 12 |
Hb_001780_030 |
0.1563577766 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 13 |
Hb_004884_170 |
0.1564815064 |
- |
- |
hypothetical protein AALP_AA1G067000 [Arabis alpina] |
| 14 |
Hb_004007_130 |
0.1575035507 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 15 |
Hb_004968_040 |
0.1614993635 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 16 |
Hb_000567_290 |
0.1630775714 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 17 |
Hb_001514_170 |
0.1642897867 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 18 |
Hb_001584_360 |
0.1650067952 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |
| 19 |
Hb_000431_100 |
0.1667802574 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 20 |
Hb_001328_070 |
0.1671985609 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |