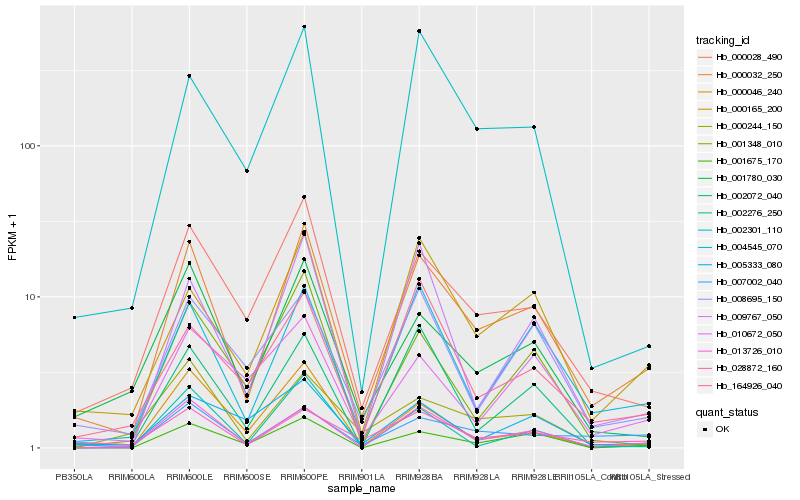
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_250 |
0.0 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 2 |
Hb_013726_010 |
0.1314317968 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000028_490 |
0.1445732334 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 4 |
Hb_004545_070 |
0.1489687361 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_001675_170 |
0.1559261162 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 [Malus domestica] |
| 6 |
Hb_000165_200 |
0.1631516485 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_028872_160 |
0.1640230084 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |
| 8 |
Hb_001780_030 |
0.1655811976 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 9 |
Hb_007002_040 |
0.175648696 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 10 |
Hb_008695_150 |
0.1866816005 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009767_050 |
0.1878852187 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000046_240 |
0.192074801 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 13 |
Hb_002301_110 |
0.1924805127 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 14 |
Hb_164926_040 |
0.1949172222 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 15 |
Hb_001348_010 |
0.1961886661 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 16 |
Hb_000244_150 |
0.1972911442 |
- |
- |
PREDICTED: chromatin assembly factor 1 subunit FAS2 [Jatropha curcas] |
| 17 |
Hb_002072_040 |
0.1972936623 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 18 |
Hb_010672_050 |
0.1973974974 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 19 |
Hb_002276_250 |
0.1976288566 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005333_080 |
0.1978809466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |