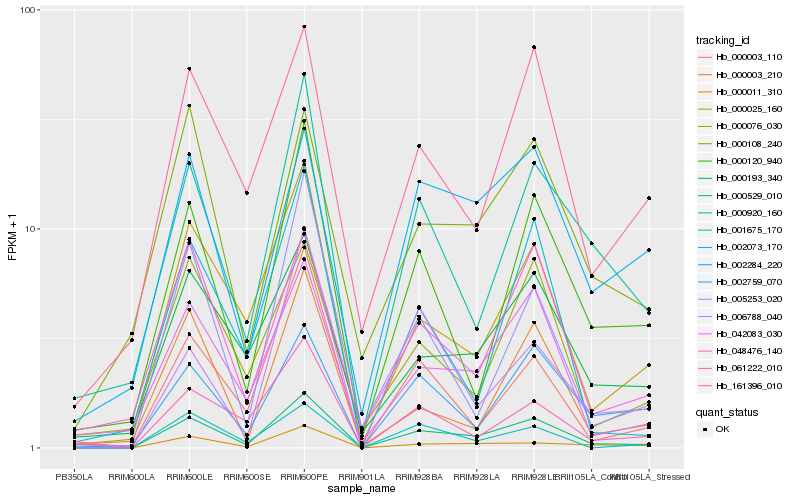
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_010 |
0.0 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 2 |
Hb_000025_160 |
0.1484119748 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 3 |
Hb_042083_030 |
0.1490593772 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000011_310 |
0.1577787831 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 5 |
Hb_000920_160 |
0.1629919767 |
- |
- |
annexin [Manihot esculenta] |
| 6 |
Hb_000120_940 |
0.1638511837 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_001675_170 |
0.1661503791 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 [Malus domestica] |
| 8 |
Hb_002759_070 |
0.167879407 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 9 |
Hb_000193_340 |
0.1682872451 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 10 |
Hb_048476_140 |
0.1722318842 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 11 |
Hb_061222_010 |
0.1780115439 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 12 |
Hb_000003_110 |
0.1800483594 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 13 |
Hb_002073_170 |
0.181042741 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 14 |
Hb_161396_010 |
0.1814022385 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 15 |
Hb_005253_020 |
0.1850946552 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 16 |
Hb_000003_210 |
0.1862307797 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 17 |
Hb_000108_240 |
0.1873137498 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 18 |
Hb_000076_030 |
0.1889848432 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002284_220 |
0.1908990607 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 20 |
Hb_006788_040 |
0.1950201316 |
- |
- |
calmodulin, putative [Ricinus communis] |