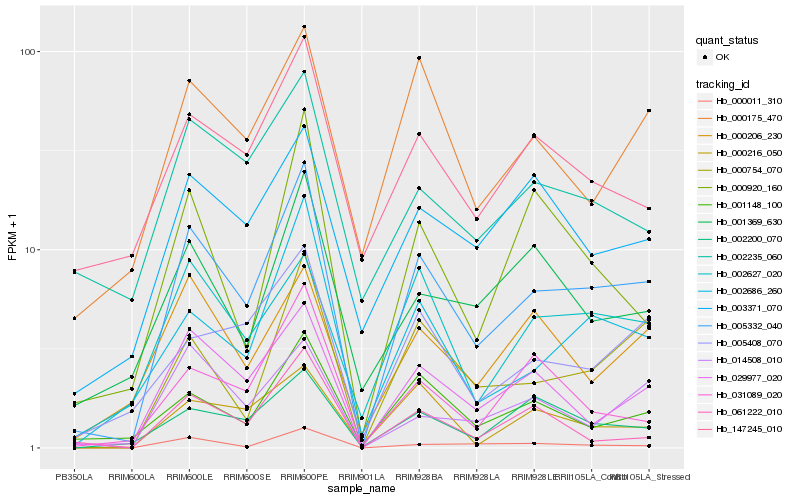
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005332_040 |
0.0 |
- |
- |
hypothetical protein RCOM_0530890 [Ricinus communis] |
| 2 |
Hb_029977_020 |
0.1413851329 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002686_260 |
0.1622963379 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 4 |
Hb_000754_070 |
0.1623431662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001148_100 |
0.1639953745 |
- |
- |
- |
| 6 |
Hb_002200_070 |
0.1747008794 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 7 |
Hb_005408_070 |
0.177028152 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 8 |
Hb_000011_310 |
0.1812514625 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 9 |
Hb_000175_470 |
0.1860263107 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 10 |
Hb_000216_050 |
0.189483568 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 11 |
Hb_002627_020 |
0.1898684726 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 12 |
Hb_014508_010 |
0.1902795309 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 13 |
Hb_031089_020 |
0.1914617121 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 14 |
Hb_000206_230 |
0.192189376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_147245_010 |
0.1934289906 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 16 |
Hb_003371_070 |
0.1937889273 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000920_160 |
0.1945215612 |
- |
- |
annexin [Manihot esculenta] |
| 18 |
Hb_061222_010 |
0.1951616688 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 19 |
Hb_001369_630 |
0.1958787159 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 20 |
Hb_002235_060 |
0.1978547695 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |