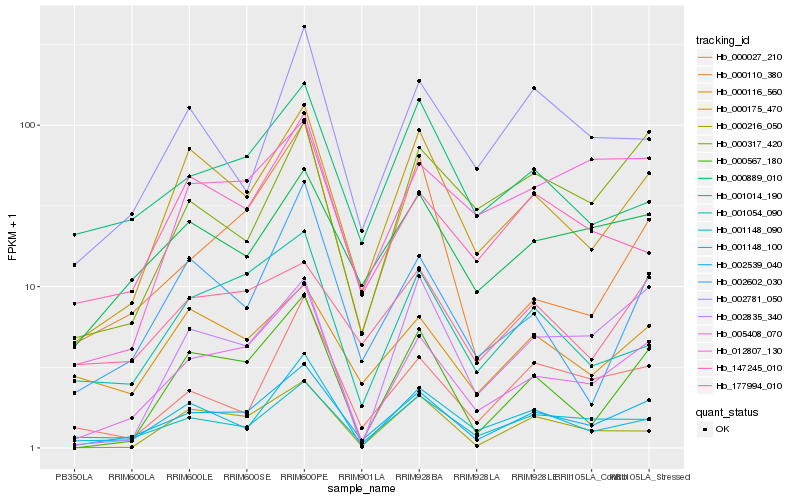
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002539_040 |
0.0 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 2 |
Hb_005408_070 |
0.0893914307 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 3 |
Hb_000175_470 |
0.1157339201 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 4 |
Hb_001148_090 |
0.1418825818 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 5 |
Hb_000567_180 |
0.1469608317 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 6 |
Hb_000889_010 |
0.154663364 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 7 |
Hb_001148_100 |
0.166539145 |
- |
- |
- |
| 8 |
Hb_000027_210 |
0.1681657937 |
- |
- |
Os01g0689300, partial [Oryza sativa Japonica Group] |
| 9 |
Hb_001054_090 |
0.1729151572 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 10 |
Hb_002781_050 |
0.1736305086 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 11 |
Hb_000116_560 |
0.174926476 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 12 |
Hb_002602_030 |
0.1751713577 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 13 |
Hb_177994_010 |
0.1787619821 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 14 |
Hb_002835_340 |
0.1813533782 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 15 |
Hb_000317_420 |
0.181838258 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 16 |
Hb_001014_190 |
0.1818923008 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 17 |
Hb_012807_130 |
0.1821451336 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 18 |
Hb_000110_380 |
0.1839090635 |
- |
- |
PREDICTED: uncharacterized protein LOC105642019 [Jatropha curcas] |
| 19 |
Hb_147245_010 |
0.1845532865 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 20 |
Hb_000216_050 |
0.1854683806 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |