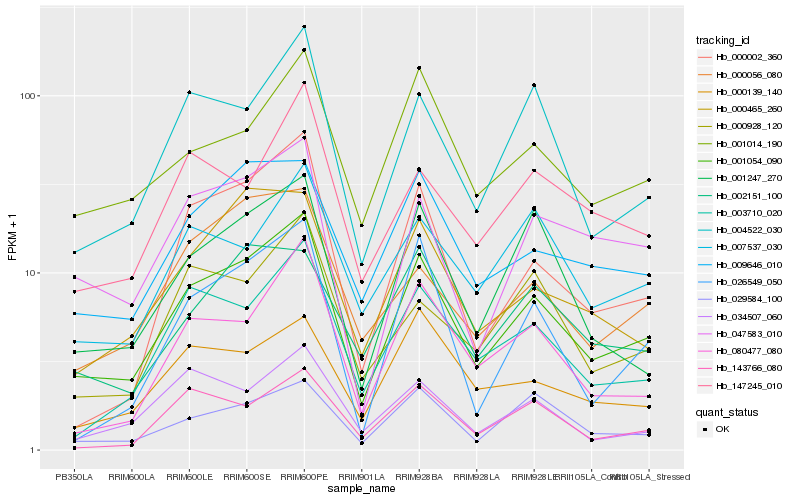
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001054_090 |
0.0 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 2 |
Hb_004522_030 |
0.1183717924 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 3 |
Hb_009646_010 |
0.1256627562 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 4 |
Hb_003710_020 |
0.1268930681 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 5 |
Hb_047583_010 |
0.1277314171 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 6 |
Hb_001014_190 |
0.1289591516 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 7 |
Hb_080477_080 |
0.1303381794 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 8 |
Hb_029584_100 |
0.1330153715 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 9 |
Hb_001247_270 |
0.1356607655 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000002_360 |
0.1365729071 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 11 |
Hb_000465_260 |
0.137838143 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 12 |
Hb_000056_080 |
0.143157674 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000928_120 |
0.1438093494 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 14 |
Hb_007537_030 |
0.1451272776 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_147245_010 |
0.1458838731 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 16 |
Hb_000139_140 |
0.1469953233 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 17 |
Hb_034507_060 |
0.1470317015 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 18 |
Hb_143766_080 |
0.1492567512 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002151_100 |
0.149368942 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 20 |
Hb_026549_050 |
0.1494902583 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |