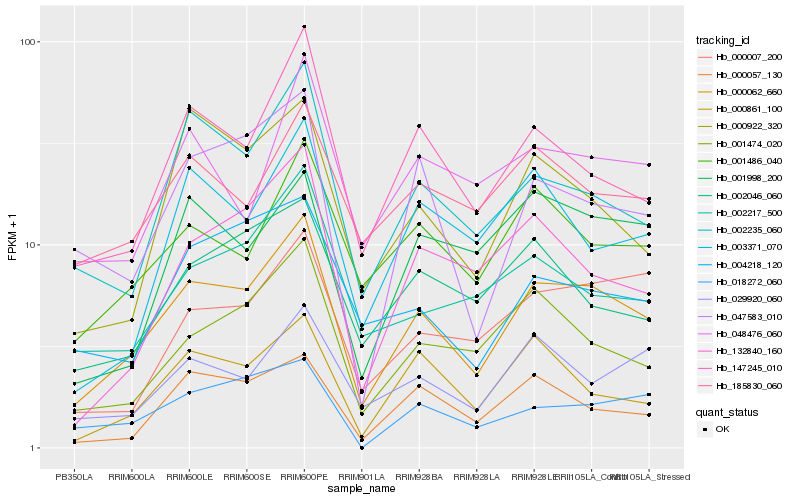
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_660 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 2 |
Hb_147245_010 |
0.1306543448 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 3 |
Hb_001486_040 |
0.1384831022 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002235_060 |
0.1398714931 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_001474_020 |
0.1405810806 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 6 |
Hb_000861_100 |
0.1489318188 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000057_130 |
0.1493096802 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 8 |
Hb_185830_060 |
0.1506811913 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 9 |
Hb_029920_060 |
0.1510894042 |
- |
- |
PREDICTED: UPF0554 protein-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_047583_010 |
0.1518422103 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 11 |
Hb_002217_500 |
0.1532605409 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 12 |
Hb_000007_200 |
0.1541821277 |
- |
- |
PREDICTED: choline/ethanolaminephosphotransferase 1 [Jatropha curcas] |
| 13 |
Hb_132840_160 |
0.1556226673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_018272_060 |
0.1558804638 |
- |
- |
PREDICTED: mavicyanin-like [Citrus sinensis] |
| 15 |
Hb_003371_070 |
0.1563204302 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_048476_060 |
0.1564559551 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002046_060 |
0.1564652397 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 18 |
Hb_000922_320 |
0.15693802 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 19 |
Hb_004218_120 |
0.1585736715 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001998_200 |
0.1603900046 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |