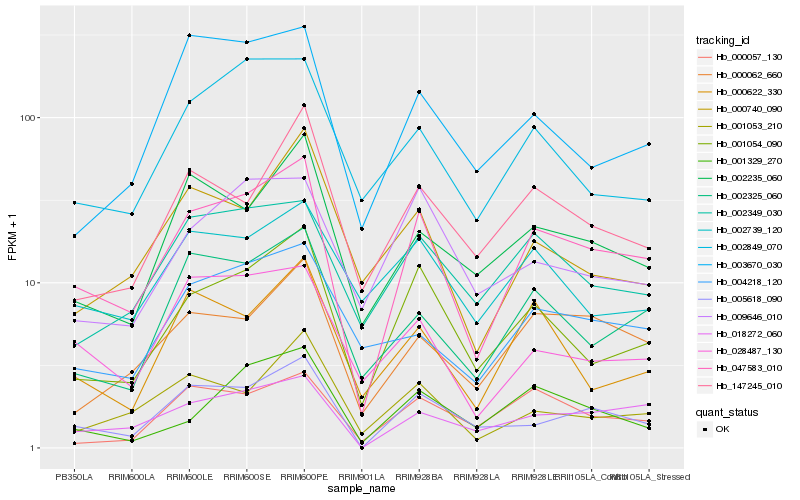
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_047583_010 |
0.0 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 2 |
Hb_001054_090 |
0.1277314171 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 3 |
Hb_018272_060 |
0.1336787037 |
- |
- |
PREDICTED: mavicyanin-like [Citrus sinensis] |
| 4 |
Hb_005618_090 |
0.1363034671 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA20, putative [Ricinus communis] |
| 5 |
Hb_002325_060 |
0.1512598038 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 6 |
Hb_000062_660 |
0.1518422103 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 7 |
Hb_002235_060 |
0.1626408527 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_002349_030 |
0.1631384921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_028487_130 |
0.1644295894 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 10 |
Hb_001053_210 |
0.1660528348 |
- |
- |
- |
| 11 |
Hb_000057_130 |
0.167081152 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 12 |
Hb_147245_010 |
0.1691700695 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 13 |
Hb_000622_330 |
0.1692488581 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 14 |
Hb_004218_120 |
0.1693244237 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002849_070 |
0.1710393525 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001329_270 |
0.1767116651 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 17 |
Hb_000740_090 |
0.1775198408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003670_030 |
0.1775820204 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 19 |
Hb_009646_010 |
0.1782013114 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 20 |
Hb_002739_120 |
0.1787465159 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |